Abstract
In this study, we provide the first report of the complete mitochondrial genome of Emydocephalus ijimae. The mitogenome length is 18,259 bp and includes 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and three non-coding regions. The sequence presented could be very useful for further phylogenetic and evolutionary study.
Sea snakes, a representative marine reptile group in the family Elapidae, comprise 63 Hydrophiinae species (true sea snakes) and eight Laticaudinae species (sea kraits) (Reptile Database Citation2019). Although the complete mitochondrial genome of three sea kraits is known (Kim et al. Citation2018), there are no published studies for any species of the subfamily Hydrophiinae.
To investigate the phylogenetic relationships of the three subfamilies of Elapidae snakes, we determined the complete mitochondrial genome of Emydocephalus ijimae (Stejneger 1898). Although genus Emydocephalus is not clearly distinguished morphologically (Rasmussen and Ineich Citation2010; Lukoschek & Sanders Citation2010), genetic analysis results for additional analysis are extremely rare. The specimen of E. ijimae was collected from the port in Serakaki, Onna, Kunigami District, Okinawa, Japan (26°30′18.24″ N and 127°52′48.37″ E) on August 05, 2013. The specimen was preserved in 70% ethanol and kept at the National Marine Biodiversity Institute of Korea (Voucher No. MABIK AR00000052).
We extracted the whole genomic DNA from the muscle tissue of the specimen using the phenol-chloroform isoamyl alcohol method as described by Asahida et al. (Citation1996) and proteinase K (Bioneer, Daejeon, South Korea). We designed sixteen primer sets to amplify the segments by long range PCR. The whole mitochondrial DNA was sequenced using the Sanger method using a 3730xl DNA analyzer (The Applied Biosystems, CA, USA). Sequencing was performed at Macrogen Inc. (Seoul, South Korea). To assemble and annotate the mitochondrial DNA sequence, we used Geneious 9.0.4 (Biomatters Ltd., Auckland, New Zealand), tRNA Scan-SE1.21 (http://lowelab.ucsc.edu/tRNA Scan-SE/) (Lowe and Eddy Citation1997), and Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004). The phylogenetic tree for E. ijimae and other related elapid snakes was constructed with PAUP* v4.0b10 (Swofford Citation2003) using the 13 protein-coding genes obtained in this study and from GenBank (). We applied the Bayesian inference and maximum likelihood method to construct the tree with 1,000 bootstrap replicates.
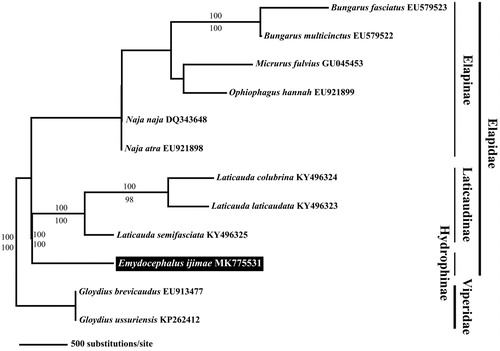
Figure 1. Maximum-likelihood (ML) tree based on the 13 mitochondrial protein-coding genes of Emydocephalus ijimae with other nine elapid snakes. We used each two species in Viperidae as the outgroup. The accession number of the mitogenomes, which obtained from GenBank, indicated after the scientific name of each species. On each branch, Bayesian posterior probabilities (above) and bootstrap value (below) are denoted.
The total length of the E. ijimae mitochondrial genome was 18,259 bp (GenBank Accession No. MK775531), with a base composition of 33.1% A, 27.6% T, 12.6% G, and 26.8% C, which showed an A-T rich (60.7%) feature. The mitogenome comprised 13 protein-coding genes, 2 rRNA genes (12S and 16S rRNA), 22 tRNA genes, and 3 non-coding regions of two control regions and an L-strand replication origin. The arrangement pattern and transcribing directions of the mitogenome were identical to those of other elapid snakes (three laticaudini species and Naja Naja), which were previously reported (Yan et al. Citation2008; Kim et al. Citation2018).
The phylogenetic tree for E. ijimae and other related elapid snakes shows that true sea snakes and other elapid snakes including sea kraits (Genus Laticauda) form a sister group (). Additionally, this result is similar to the previous result by Slowinski and Keogh (Citation2000) regarding the phylogenetic relationships of elapid snakes based on the cytochrome b sequence and that of Keogh (Citation1998) regarding the phylogeny of elapid snakes based on 16S rRNA and cytochrome b.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Asahida T, Kobayashi T, Saitoh K, Nakayama I. 1996. Tissue preservation and total DNA extraction form fish stored at ambient temperature using buffers containing high concentration of urea. Fisheries Sci. 62:727–730.
- Keogh JS. 1998. Molecular phylogeny of elapid snakes and a consideration of their biogeographic history. Biol J Linn Soc. 63:177–203.
- Kim IH, Park J, Suk HY, Bae HG, Min MS, Tsai TS, Park D. 2018. Phylogenetic relationships of three representative sea krait species (genus Laticauda; elapidae; serpentes) based on 13 mitochondrial genes. Mitochondrial DNA A. 29:772–777.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Lukoschek V, Sanders K. 2010. Emydocephalus ijimae. The IUCN Red List of Threatened Species. 2010:e.T176706A7286976.
- Rasmussen AR, Ineich I. 2010. Species Diversity in the Genus Emydocephalus Krefft, 1869 (Serpentes, Elapidae, Hydrophiinae): Insight from Morphology and Anatomy. Herpetol Rev. 41:285–290.
- Reptile Database. 2019. The Reptile database, URL: http://www.reptile-database.org (accessed on Fab 1, 2019).
- Slowinski JB, Keogh JS. 2000. Phylogenetic relationships of elapid snakes based on cytochrome b mtDNA sequences. Mol Phylogenet Evol. 15:157–164.
- Swofford DL. 2003. PAUP*: Phylogenetic Analysis Using Parsimony, Version 4.0 b10.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yan J, Li H, Zhou K. 2008. Evolution of the mitochondrial genome in snakes: gene rearrangements and phylogenetic relationships. BMC Genomics. 9:569.
