Abstract
Uvaria macrophylla (Annonaceae) is an erect shrub with multiple medicinal properties. In this study, we report the complete chloroplast genome of U. macrophylla, assembled from whole-genome high-throughput sequencing reads, as a resource for future studies on the phylogeny and evolution of Annonaceae. The chloroplast genome was 192,782 bp in length, with a large single-copy (LSC) region of 83,581 bp, a small single-copy (SSC) region of 3,741 bp, separated by two inverted repeat (IR) regions of 52,730 bp each. It was predicted to contain 151 genes, with an overall GC content of 38.7%. Phylogenetic analysis of 105 protein-coding sequences of 13 plant plastomes showed that U. macrophylla is closest to Annona cherimola.
Uvaria macrophylla, an erect shrub, belongs to the family Annonaceae. U. macrophylla is widely distributed in Hainan, Guangdong and Guangxi provinces of southern China, occurring in low altitude boscage and woodland (Wu et al. Citation2004). The medicinal properties of U. macrophylla has been widely studied by scientists (Zafra Polo et al. Citation1998; Wang et al. Citation2005; Zhu et al. Citation2011; Chen et al. Citation2012). Some studies revealed that the constituents in the extracts from U. macrophylla exhibited inhibitory activities toward a number of human cancer cell lines (Wang et al. Citation2002, Citation2003; Zhang et al. Citation2002). In this study, we characterized the complete chloroplast genome sequence of U. macrophylla as a resource for future genetic studies on this plant and its related species.
Leaf samples of U. macrophylla were obtained from the Guangzhou Institute of Forestry and Landscape Architecture, with voucher specimen (SYS-Bore-2019-04) being deposited at Sun Yat-sen University Herbarium (SYS). After DNA extraction, a ∼350-bp insertion library was constructed and then sequenced (paired-end 150 bp) on the Illumina Hiseq Xten platform, generating approximately 4 Gb raw data. Using a partial rbcL gene from U. macrophylla (KP094324) as a seed, all data were assembled into an intact circular chloroplast genome using the software NOVO Plasty (Dierckxsens et al. Citation2017). The assembled chloroplast genome sequence was annotated using DOGMA (Wyman et al. Citation2004) and manually corrected.
The complete chloroplast genome sequence of U. macrophylla (GenBank accession MH992130) was 192,782 bp in length, with a large single-copy (LSC) region of 83,581 bp, a small single-copy (SSC) region of 3,741 bp, and two inverted repeat (IR) regions of 52,730 bp each. A total of 151 genes were predicted, consisting of 105 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The overall GC content was 38.7%.
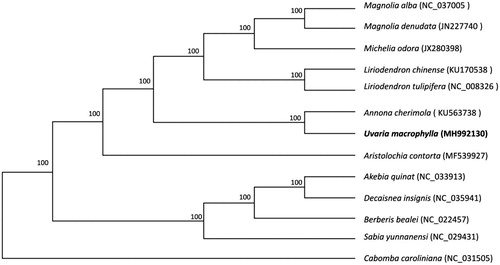
To understand the phylogenetic position of U. macrophylla, an ML tree was drawn for U. macrophylla and 12 other representative species from Magnoliales, Ranales, Sapindales and Aristolochiales. The homologous blocks in the chloroplast genomes among these 13 species were extracted using HomBlocks tool (Bi et al. Citation2018). These blocks were then aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and RaxML (Stamatakis, Citation2014) was used to construct a maximum likelihood tree with C. caroliniana as outgroup. As shown in , U. macrophylla is closest to Annona cherimola from the same family.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. A multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Chen Z, Liu XR, Liu YL, et al. 2012. ChemInform Abstract: Two New Flavones from Uvaria macrophylla Roxb. var. microcarpa and Their Cytotoxic Activities. Cheminform. 43:2857–2863.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang S, Zhang PC, Chen RY, et al. 2002. A Novel Dihydro flavone from the Roots of Uvaria Macrophylla[J]. Chinese Chem Lett. 13:857–858.
- Wang S, Chen RY, Yu SS, Yu DQ. 2003. Uvamalols D-G: novel polyoxygenated seco-cyclohexenes from the roots of Uvaria macrophylla. J Asian Nat Prod Res. 5:17–23.
- Wang S, Zhang PC, Chen RY, Dai SJ, Yu SS, Yu DQ. 2005. Four new compounds from the roots of Uvaria macrophylla. J Asian Nat Prod Res. 7:687–694.
- Wu ZY, Raven P, Hong DY, et al. 2004. Flora Reipublicae Popularis Sinicae. Beijing (China): Science Press (in Chinese), p. 126.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zafra Polo MC, Figadère B, Gallardo T, et al. 1998. Natural acetogenins from Annonaceae, synthesis and mechanisms of action. Phytochemistry. 48:1087–1117.
- Zhang HL, Wang S, Chen RY, Yu DQ. 2002. Studies on chemical constituents of Uvaria macrophylia. Yao Xue Xue Bao. 37:124–127.
- Zhu XY, Lin SW, Lu RM. 2011. Analysis of chemical components of volatile oils from leaves of Uvaria microcarpa Champ.ex Benth by SFE-CO2. Medicinal Plant. 39:13376–13377.