Abstract
Blue-winged Minla is a small-sized bird, belonging to the Family Leiothrichidae of the Passeriformes. The population is suspected to be in decline owing to ongoing habitat destruction and fragmentation. Therefore, we sequenced and analyzed the Blue-winged Minla mitochondrial genome to provide a theoretical basis for protecting the brids. The mitochondrial genome of Blue-winged Minla was 17862 bp, it contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 2 control regions. The overall base composition of the mitochondrial was 30.88% A, 13.82% G, 23.86% T, 31.44% C.
Blue-winged Minla (Minla cyanouroptera) is a small-sized bird, belonging to the Family Leiothrichidae of the Passeriformes. The length of body is 13–16 cm, similar in gender, grey-brown overhead with light blue and black stripes. Blue-winged Minla natural habitat is subtropical or tropical moist montane forests and it is distributed in Bhutan, Cambodia, China, India, Lao People’s Democratic Republic, Nepal, Thailand, Viet Nam. Despite the fact that the population trend appears to be decreasing, the decline is not believed to be sufficient rapid to approach the thresholds for Vulnerable under the population trend criterion. Therefore, the species is evaluated as Least Concern (BirdLife International Citation2016). The population is suspected to be in decline owing to ongoing habitat destruction and fragmentation. Therefore, we sequenced and analyzed the Blue-winged Minla mitochondrial genome to provide a theoretical basis for protecting the brids.
The sample was collected from Ya’an (30°0′47.20″N, 103°02′25.51″E) Sichuan province of China and Stored at the Sichuan Agricultural University Museum, China and sample ID was 9092.xc Total DNA of Blue-winged Minla was isolated from muscle tissue by the traditional phenol-chloroform method (Sambrook Citation2001). Fifteen pairs of primers were used to amplify the overlapping segment of its mitochondrial genomic by the Polymerase Chain Reaction (PCR). Then splicing sequences and analyzed complete mitochondrial genome by software DNAstar (Burland Citation2000). We have submitted the complete mitochondrial sequences of Blue-winged Minla to GeneBank and got the accession number: MK779708.
The complete mitochondrial genome of Blue-winged Minla published in this paper, and mitochondrial DNA is 17862 base pairs in length, contained 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 2 control regions (Dloop1, Dloop2) which was similar to other mitochondrial genome of Passeriformes (Xue et al. Citation2016; Liu et al. Citation2018). The overall base composition of the genome was 30.88% A, 13.82% G, 23.86% T, 31.44% C. All tRNA genes possess the typical clover leaf secondary structure except for tRNAser which lack arm of dihydrouracil (DHU). NADH6 and eight tRNA genes were transcribed from the L-strand, the rest genes of Blue-winged Minla were encoded on the H-strand. All protein-coding genes used ATG as start coding, except for NADH6 which began with CTA, there are two types stop codon which complete or incomplete, 6 PCGs (NADH1, COXII, ATPase8, ATPase6, NADH4L, CYTB) shared stop codon TAA, 2 PCGs (NADH2, NADH6) shared stop codon CAT, COXI and NADH5 stopped with AGG, AGA, 3 PCGs shared incomplete codon. Consistent with previous research results, two ribosomal RNA genes were located between the tRNAphe and tRNALeu which separated by the tRNAVal (Zhang Citation2015; Weiqiang He et al. Citation2018).
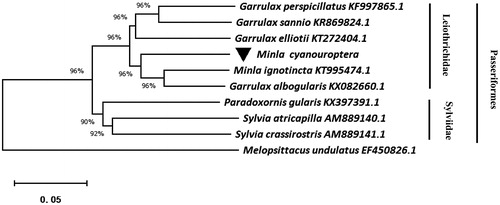
We downloaded eight whole mitochondrial sequence of Passeriformes from the NCBI, and Melopsittacus undulates was used as the outgroup (). Using the MEGA10.0 for neighbor-joining (NJ) methods to construct a phylogenetic tree (Kumar et al. Citation2018). The phylogenetic tree suggested that nine species clustered into two clades, including Leiothrichidae and Sylviidae. Other than this, we found that Mina cyanouroptera and Mina ignotincta have close relationship.
Acknowledgements
We thank the Qian Su, Xue Liu, Pu Zhao, Yuhan Wu, Tianrui Xia, Cong Wang, Kechu Zhang for their assistance in experiments and data analysis.
Disclosure statement
The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
Additional information
Funding
References
- BirdLife International. 2016. Siva cyanouroptera. The IUCN Red List of Threatened Species 2016:e.T22716576A94501013.http://dx.doi.org/10.2305/IUCN.UK.2016-3. RLTS. T22716576A94501013.en. Downloaded on 4 April 2019.
- Burland TG. 2000. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Liu X, Xu H, Zhou Y, Li D, Ni Q, Zhang M, Xie M, Wen A, Wang Q, Wu J, et al. 2018. Complete characteristics and phylogenetic relationships of the Garrulax albogularis mitochondrial genome (Passeriformes: Timaliidae). Mitochondrial DNA Part B. 3:1272–1273.
- Sambrook JR. 2001. Molecular cloning: a laboratory manual, 3rd edn. New York (NY): Cold Spring Harbor Laboratory Press, pp 12–72.
- Weiqiang He HX, Li D, Xie M, Zhang M, Ni Q, Wu J, Wen A, Wang Q, Zhu G, Yao Y. 2018. Complete mitochondrial genome and phylogenetic analysis of Black-chinned Yuhina (Yuhina nigrimenta). Mitochondrial DNA Part B. 4:605–606.
- Xue H, Zhang H, Li Y, Wu X, Yan P, Wu XB. 2016. The complete mitochondrial genome of Garrulax cineraceus (Aves, Passeriformes, Timaliidae). Mitochondrial DNA PartA. 27:147–148.
- Zhang H, Li Y, Wu X, Xue H, Yan P, Wu X-B. 2015. The complete mitochondrial genome of Paradoxornis webbianus (Passeriformes, Muscicapidae). Mitochondrial DNA PartB. 26:879–880.