Abstract
The nearly complete sequence of the mitochondrial DNA of Calineuria stigmatica has been completed and annotated in this study. The circular genome is 15,070 bp in length with an A + T content of 61.8% and contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and two ribosomal RNA (rRNA) genes. The control region can only be assembled partially. All PCGs use normal start codon ATN, while COI, ND1, and ND5 use CCG, TTG, and GTG as start codon, respectively. Meanwhile, 10 PCGs use the typical termination codons TAN, except COII, ND4, ND5, which stopped with the incomplete terminaton signal T––. Based on 13 PCGs and two rRNAs using the Bayesian (BI) method supported that C. stigmatica was closely grouped with four other Acroneuriinae species. Our results provide basic data for further study of phylogeny in Plecoptera.
Calineuria Ricker, 1954, a subgenus of Acroneyria Pictet, 1841, was later elevated to genera by Illies (Citation1965). Uchida (Citation1983) transferred the species A. stigmatica to the genus Calineuria. It is a small genus currently including seven species and is known from the Nearctic, eastern Palearctic, and Oriental regions, and Calineuria stigmatica is considered rare in the stonefly groups which only distributed in Japan (Uchida Citation1983; DeWalt et al. Citation2019). Mitochondrial DNA is a powerful molecular tool for genetic research and has proven to be useful for rapid identification and genetic source determination (Cameron Citation2014). To date, nine perlid but only four species from Acroneuriinae have been published previously, and the genus Calineuria has not yet been involved (Cao, Li, et al. Citation2019; Cao, Wang, et al. Citation2019; Elbrecht et al. Citation2015; Huang et al. Citation2015; Li et al. Citation2019; Qian et al. Citation2014; Wang, Ding, et al. Citation2016; Wang, Wang, et al. Citation2016). In this study, we characterized the mitochondrial genome of C. stigmatica by high-throughput sequencing for the first time. This study would supply important basis for conservation genetics of C. stigmatica, and promote the phylogenetic analysis of Plecoptera.
The male adult sample of C. stigmatica was collected from Yamagata Prefecture (37.951°N, 139.698°E), Japan in 2015 by Xingyue Liu. The thorax muscle of the specimen was used to extract total genomic DNA. Vouchers consisting of the remaining stoneflies (No. Vhl-0064) were deposited in the Entomological Museum of Henan Institute of Science and Technology (HIST), Henan Province, China.
The annotated mitogenome of C. stigmatica is 15,070 bp in length, which has been deposited into GenBank with the accession number MG677941. It encoded 37 genes as in other insect mitogenomes, including 13 PCGs, 22 tRNA genes, two rRNA genes, and a partial control region. It had a base composition of A 33.9%, T 27.9%, C 25.2%, and G 13.0%. The A + T content of PCGs, tRNAs, and rRNAs was 60.1, 67.0, and 67.5%, respectively. All PCGs use ATG as start codon except the ND1 and ND5 genes used TTG and GTG. Ten PCGs used the typical termination codons TAA or TAG, whereas only three PCGs (COII, ND4, and ND5) stop with the signal T––. The 22 tRNA genes size varied from 64 to 72 bp, comprising a total length of 1475 bp. In addition to the control region, there were 44 nucleotides dispersed in eight intergenic spacers, ranging from 1 to 16. Gene overlaps were also found at sixteen gene junctions involving 44 nucleotides with the longest overlap (eight nucleotides) between tRNATrp and tRNACys.
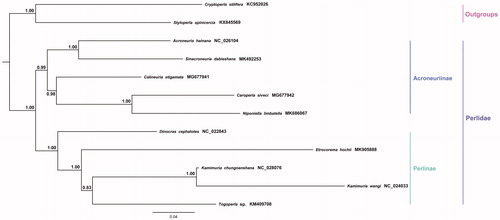
The phylogenetic relationship of C. stigmatica was constructed with the sequences of 13 PCGs and two rRNAs from 10 perlid stoneflies (including an unpublished species Etrocorema hochii) and two species as outgroups by MEGA 7.0 (Kumar et al. Citation2016) using the Bayesian (BI) method to conduct the topology tree (). Our result showed that C. stigmatica was closely grouped with four other Acroneuriinae species, which is in accordance with the traditional morphological classification. More mitogenome information of Perlidae species is needed for the further phylogenetic studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Cao JJ, Li WH, Wang Y. 2019. The complete mitochondrial genome of a Chinese endemic stonefly species, Sinacroneuria dabieshana (Plecoptera: Perlidae). Mitochondrial DNA Part B Resour. 4:1327–1328.
- Cao JJ, Wang Y, Li N, Li WH, Chen XL. 2019. Characterization of the nearly complete mitochondrial genome of a Chinese endemic stonefly species, Caroperla siveci (Plecoptera: Perlidae). Mitochondrial DNA Part B Resour. 4:553–554.
- DeWalt RE, Maehr MD, Neu-Becker U, Stueber G. 2019. Plecoptera species file online. [accessed 2019 Jun 26]. http://Plecoptera.SpeciesFile.org.
- Elbrecht V, Poettker L, John U, Leese F. 2015. The complete mitochondrial genome of the stonefly Dinocras cephalotes (Plecoptera, Perlidae). Mitochondrial DNA. 26:469–470.
- Huang MC, Wang YY, Liu XY, Li WH, Kang ZH, Wang K, Li XK, Yang D. 2015. The complete mitochondrial genome and its remarkable secondary structure for a stonefly Acroneuria hainana Wu (Insecta: Plecoptera, Perlidae). Gene. 557:52–60.
- Illies J. 1965. Phylogeny and zoogeography of the Plecoptera. Annu Rev Entomol. 10:117–140.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li JF, Cao JJ, Wang Y, Kong FB. 2019. The mitochondrial genome analysis of a stonefly, Niponiella limbatella (Plecoptera: Perlidae). Mitochondrial DNA B Resour. 4:1666–1667.
- Qian YH, Wu HY, Ji XY, Yu WW, Du YZ. 2014. Mitochondrial genome of the stonefly Kamimuria wangi (Plecoptera: Perlidae) and phylogenetic position of Plecoptera based on mitogenomes. PLoS One. 9:e86328.
- Uchida S. 1983. A new species of Calineuria (Plecoptera, Perlidae) from Japan, with notes on the Japanese species of the genus. Jpn J Entomol. 51:622–627.
- Wang K, Ding SM, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Kamimuria chungnanshana Wu, 1948 (Plecoptera: Perlidae). Mitochondrial DNA A. 27:3810–3811.
- Wang K, Wang YY, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Togoperla sp. (Plecoptera: Perlidae). Mitochondrial DNA A. 27:1703–1704.