Abstract
Rumex crispus has high medicinal value which belongs to the family Polygonaceae. We sequenced the complete chloroplast genome of R. crispus, which is 158,851 bp in length. A total of 111 unique genes have been predicted in the chloroplast genome of this species, consisting of 77 protein-coding sequences, 30 tRNA and 4 rRNA genes. A maximum likelihood (ML) phylogenetic tree based on 80 protein-coding genes of 18 Polygonaceae species showed the phylogenetic position of R. crispus within the family Polygonaceae. These results facilitate population and biological studies of R. crispus and benefit further investigations of this important species.
Rumex crispus is a perennial plant and belongs to the family Polygonaceae. R. crispus has a long history of domestic herbal use because of its high medicinal values. The roots, leaves and seeds of R. crispus have been reported to possess broad curative activities, including antioxidant, antimicrobial (Kim et al. Citation1999; Yildirim et al. Citation2001), hepatoprotective, anti-inflammatory and analgesic (Lee et al. Citation2007). Nevertheless, despite its importance, few studies have described the genome of R. crispus. Therefore, we establish and characterize the organization of the complete chloroplast genome of R. crispus. The results will also provide genetic information of this species as well as valuable knowledge for future research in the identification of Polygonaceae species.
Dried radix and rhizome powder of R. crispus (batch number 121291-201102) were collected from National Institutes for Food and Drug Control. Meanwhile, the specimens (voucher numbers TDH-2) were deposited in Tianjin State Key Laboratory of Modern Chinese Medicine, Tianjin, China. Total genomic DNA was extracted using Extract Genomic DNA Kit with a standard protocol. The DNA was sequenced using the Illumina Hiseq X Ten platform with 800 bp insert size. The clean data was de novo assembled using NOVOPlasty v3.1 (Dierckxsens et al. Citation2017) and GetOrganelle pipeline (Jin et al. Citation2018). Then, the resulting contigs were identified and rearranged by mapping to reference genome Rumex acetosa (Gui et al. Citation2018) (Accession number: MH359405). Annotation of the chloroplast genome was performed using CpGAVAS2 (Shi et al. Citation2019) and GeSeq (Tillich et al. Citation2017). Subsequently, the annotation of two methods above was compared and inspected manually. The tRNAscan-SE search server (Lowe and Eddy Citation1997) was employed to verify the annotations of tRNA genes. Finally, the physical genome map was illustrated with OGDraw (Greiner et al. Citation2019), and the complete annotated plastome of R. crispus was deposited into GenBank (accession numbers: MN055629).
In this study, the complete chloroplast genome presented 158,851 bp in size, with a total of 111 unique genes including 77 protein-coding sequences, 30 tRNA and 4 rRNA genes. Among these genes, 15 genes (rpl16, rpl2, rps16, rpoC1, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, ndhA, ndhB, petB, petD, and atpF) contained one intron and two genes (clpP and ycf3) harbored two introns.
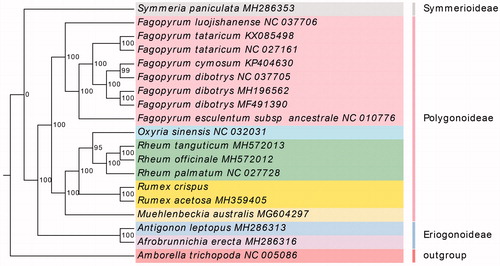
To infer the phylogenetic position of R. crispus within the family Polygonaceae, 17 Polygonaceae species plastomes were downloaded from the Genbank database. All 80 protein-coding genes were extracted and aligned using MAFFT v7 (Katoh and Standley Citation2013) with default parameters and concatenated to a final data matrix. RAxML v8 (Stamatakis Citation2014) was employed to reconstruct the maximum-likelihood (ML) phylogeny based on the GTRGAMMA model using 1000 replicates of bootstrap analysis. In the maximum-likelihood (ML) tree, most of the nodes had a 100% bootstrap value. Rumex exhibited the closest relationship with other two genera, Rheum and Oxyria. Meanwhile, the phylogenetic positions of Rumex were in agreement with recent studies (Gao et al. Citation2019) (). Overall, the results of this study will help to provide valuable resources for molecular biology and evolutionary studies of R. crispus.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Gao H, Tan W, Yang T, Tian X. 2019. The complete chloroplast genome of Rheum rhabarbarum. Mitochondrial DNA B. 4:1965–1966.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47:W59–W64.
- Gui L, Jiang S, Wang H, Nong D, Liu Y. 2018. Characterization of the complete chloroplast genome of sorrel (Rumex acetosa). Mitochondrial DNA B. 3:902–904.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Yi T-S, Li D-Z. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 30:772–780.
- Kim IH, Han T. Yunchon (Korea Republic, Yunchon Adlay Experiment Station), Chang SWHU, Chunchon (Korea Republic). Department of Biology. 1999. Anthraquinone productivity by the cultures of adventitious roots and hairy from curled dock (Rumex crispus). v. 26. Korean.
- Lee SSSU (Seoul, Republic of Korea), Kim DHSU (Seoul, Republic of Korea), Yim DSSU (Seoul, Republic of Korea), Lee SYSU (Seoul, Republic of Korea), E-mail: [email protected]. 2007. Anti-inflammatory, analgesic and hepatoprotective effect of semen of Rumex crispus v. 38. Korean.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 25:0955–0964.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47:W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England). 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Yildirim A, Mavi A, Kara AA. 2001. Determination of antioxidant and antimicrobial activities of Rumex crispus L. extracts. J Agric Food Chem. 49:4083–4089.