Abstract
In this work, we reported the complete mitochondrial genome yielded from next-generation sequencing of Corydoras sterbai. The total length of the mitochondrial genome is 16,636 bp, with the base composition of 32.70% A, 26.40% T, 26.00% C, and 14.90% G. It contains two ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and a major non-coding control region (D-loop region). The arrangement of these genes is the same as that found in the Siluriformes. All the protein initiation codons are ATG, except for cox1 that begins with GTG. The Neighbour-Joining tree built using Geneious showed that C. sterbai was clustered with other species from the genus Corydoras. It means that its external morphological feature classification is consistent with the molecular classification results.
Corydoras, one of the nine genera of the Corydoradinae, are identified by their twin rows of armour plates along the flanks and by having fewer than 10 dorsal fin rays. They are most commonly confused with the other genera in the sub-family, namely Brochis, Scleromystax, and Aspidoras. There are more than 400 species in the genus Corydoras. Corydoras sterbai, named ‘pearl mouse’ in chinese ornamental fish market, is from smaller rivers and tributaries, as well as flooded forest pools with soft, acidic waters in central Brazil and Bolivia (South America). Now, C. sterbai is a common small-size ornamental fish in the Chinese ornamental fish market.
In order to distinguish and identify the species more accurately, we determined the mitochondrial genome sequence of the ‘pearl mouse’ C. sterbai using next-generation sequencing (Zou et al. Citation2019) and carried out mitochondrial genome structure and phylogenetic analysis. The living body of ‘pearl mouse’ was collected from the Red Star Ornamental Fish Market in Changsha, Hunan Province, China (113.03 E, 28.09 N). After anesthesia with MS-222 (3-Aminobenzoic acid ethyl ester methanesulfonate), dorsal muscle tissue was collected and preserved in 99% ethanol in Museum of Hunan Agricultural University. After DNA extraction (Tissue DNA Kit D3396-02, Omega, bio-tek) and sequencing library construction (Sangon Biotech, Shanghai), paired end reads were sequenced using HiSeq XTen PE 150 of Illumina. BBduk and BLAST + were used to assess and monitor data quality. NOVOPlasty and SPAdes were used for de novo assembly. MITOS2 server and Geneious R11 (Liu et al. Citation2019; Tan et al. Citation2019) were used to predict and annotate the mitochondrial genome. Multiple alignment (ClustalW) and Geneious Tree Builder was utilized to compare differences among the mitogenomes and build phylogenetic tree.
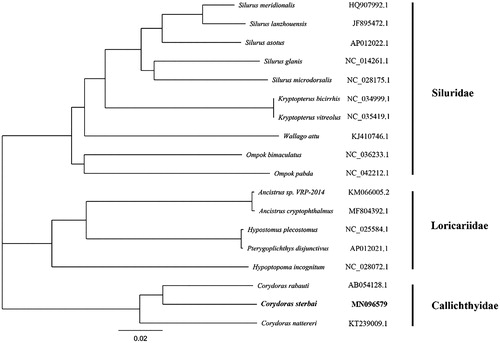
Totally 28,555,528 high-quality clean reads (150 bp PE read length) were obtained. The total length of the C. sterbai mitochondrial genome is 16,636 bp (GenBank accession number: MN096579), with the base composition of 32.70% A, 26.40% T, 26.00% C, and 14.90% G. It contains two ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and a major non-coding control region 1023 bp in length (D-loop region). The arrangement of these genes is the same as that found in the Siluriformes (Saitoh et al. Citation2003; Liu et al. Citation2019). All the protein initiation codons are ATG, except for cox1 that begins with GTG. The Neighbour-Joining tree built using Geneious with Tamura-Nei (genetic distance model) and global aligment with free end gaps (aligment type) showed that C. sterbai was clustered with other species in the genus Corydoras (). It means that its external morphological feature classification is consistent with the molecular classification results. The information of the mitogenome will be beneficial for future phylogenetic studies and specimen identification of Corydoras species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Tan J, Yin Z, Huang H, Zeng C. 2019. Characterization and phylogenetic analysis of Acheilognathus chankaensis mitochondrial genome. Mitochondrial DNA B. 4:1148–1149.
- Liu Q, Xu B, Xiao T. 2019. Complete mitochondrial genome of Corydoras duplicareus (Teleostei, Siluriformes, Callichthyidae). Mitochondrial DNA B. 4:1832–1833.
- Saitoh K, Miya M, Inoue JG, Ishiguro NB, Nishida M. 2003. Mitochondrial genomics of ostariophysan fishes: perspectives on phylogeny and biogeography. J Mol Evol. 56:464–472.
- Zou Y, Zhang J, Wu T, Wen Z. 2019. Complete mitochondrial genome of the Yunnanilus sichuanensis and phylogenetic analysis. Mitochondrial DNA B. 4:941–942.