Abstract
The complete mitochondrial genome was determined for the Cynoglossus interruptus belonging to the family Cynoglossidae. The length of the complete mitochondrial genome is 17,262 bp, consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. The gene rearrangement related to tRNAGln and a control region gene were found, forming the gene order of CR-Ile-Gln-Met. Phylogenetic analysis using mitochondrial genomes of 12 species showed that C. interruptus formed a well-supported monophyletic group with other Cynoglossus species.
The Genus Cynoglossus (Pleuronectiformes: Cynoglossidae), including about 50 species, is distributed in tropical and subtropical waters from the Atlantic coast of Africa to Western Pacific (Menon Citation1977). In the Genus Cynoglossus, six species comprising C. abbreviates, C. gracilis, C. semilaevis, C. joyneri, C. robustus, and C. interruptus were recorded from Korea (Jordan and Metz Citation1913; Mori Citation1928; Mori and Uchida Citation1934; Uchida and Yabe Citation1939; Mori Citation1952). In previous studies, Yokogawa et al. (Citation2008) reported that confirmation regarding the distribution of the C. interruptus among six species in Korean waters is required, and Kwun and Kim (Citation2016) have re-identified that the C. interruptus is distributed in Korean waters using morphological and molecular data. In this study, we first determined the complete mitochondrial genome of C. interruptus.
The C. interruptus specimen was collected from Sacheon-si, Gyeongsangnam-do, Republic of Korea (34.56 N, 127.58E). Total genomic DNA was extracted from tissue of the specimen, which has been deposited in the Marine Fish Resources Bank of Korea (MFRBK) (Voucher No. PKU56300). The mitogenome was sequenced and assembled using Illumina Hiseq 4000 sequencing platform (Illumina, San Diego, CA) and SOAP denovo assembler at Macrogen Inc. (Korea), respectively. The complete mitochondrial genome was annotated using MacClade ver. 4.08 (Maddison and Maddison Citation2005) and tRNAscan-SE 2.0 (Lowe and Chan Citation2016).
The mitochondrial genome of C. interruptus (GenBank accession no. LC482306) is 17,262 bp long, including 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. The overall base composition is 30.53% A, 24.38% C, 14.98% G, and 30.11% T, with a bias on AT content (60.64%). The 12S rRNA (943 bp) and 16S rRNA genes (1,685 bp) are located between tRNAPhe and tRNAVal and between tRNAVal and tRNALeu(UUR), respectively. Of the 13 protein-coding genes, 11 begin with an ATG start codon; the exception being the COI gene and ND3, which start with GTG and ATT, respectively. The stop codon of the protein-coding genes is TAA in ND1, COI, ATP8, ND4L, ND5, ND6 and Cytb; TAG in ND2; TA– in ATP6 and COIII; and T–– in the remaining three genes.
Additionally, gene rearrangements related to tRNA-Gln gene and control region were found. The tRNA-Gln gene has translocated from the light to the heavy strand and the control region translocated downstream to the place between ND1 and tRNA-Ile genes. Also, we detected unique gene order, CR-Ile-Gln-Met, which is different from other Cynoglossus species, forming the gene order of CR-Gln-Ile-Met (Kong et al. Citation2009; Shi et al. Citation2015; Wei et al. Citation2016; Bo et al. Citation2017; Chen et al. Citation2019).
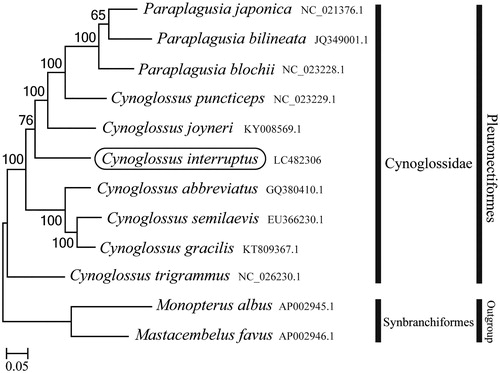
Phylogenetic trees were constructed by the maximum-likelihood method using MEGA 7.0 software (Kumar et al. Citation2016) for the newly sequenced genome and a further 11 mitochondrial genome sequences downloaded from the National Center for Biotechnology Information. We confirmed that C. interruptus formed a monophyletic group with other Cynoglossus species (). The mitogenome features of the C. interruptus provide important molecular data to confirm that C. interruptus is distributed in Korean waters.
Acknowledgements
This research was conducted using the fish specimens provided by the Marine Fish Resource Bank of Korea (MFRBK) under the Ministry of Oceans and Fisheries, Korea.
Disclosure statement
The authors report no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bo Z, Lei J, Kefeng L, Jinsheng S. 2017. The complete mitochondrial genome of Cynoglossus joyneri and its novel rearrangement. Mitochondrial DNA. 2:581–582.
- Chen S, Dong J, Luo H, Shi W, Kong X. 2019. The complete mitochondrial genome sequence of Cynoglossus roulei (Pleuronectiformes: Cynoglossidae). Mitochondrial DNA. 4:1443–1444.
- Jordan DS, Metz CW. 1913. A catalog of the fishes known from the waters of Korea. Mem Carnegie Mus. 6:1–65.
- Kong XY, Dong XL, Zhang YC, Shi W, Wang ZM, Yu ZN. 2009. A novel rearrangement in the mitochondrial genome of tongue sole, Cynoglossus semilaevis: control region translocation and a tRNA gene inversion. Genome. 52:975–984.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Kwun HJ, Kim JK. 2016. Re-identification of two tonguefishes (Pleuronectiformes) from Korea using morphological and molecular Analyses. Korean J Fish Aquat Sci. 49:208–213.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Maddison DR, Maddison WP. 2005. MacClade 4: analysis of phylogeny and character evolution. Version 4.08. Sunderland (MA): Sinauer Associates.
- Menon A. 1977. A systematic monograph of the tongue soles of the genus Cynoglossus Hamilton-Buchanan (Pisces: Cynoglossidae). Smithson Contrib Zool. 238:1–129.
- Mori T. 1928. A catalogue of the fishes of Korea. J Pan-Pacific Res Inst. 3:3–8.
- Mori T. 1952. Check list of the fishes of Korea. Mem Hyogo Univ Agric. 1:1–228.
- Mori T, Uchida K. 1934. A revised catalogue of the fishes of Korea. J Chosen Nat Hist Soc. 19:1–23.
- Shi W, Jiang JX, Miao XG, Kong XY. 2015. The complete mitochondrial genome sequence of Cynoglossus sinicus (Pleuronectiformes: Cynoglossidae). Mitochondrial DNA. 26:865–866.
- Uchida KS, Yabe H. 1939. The fish fauna of Saisyu-to (Quelpart Island) and its adjacent waters. J Chosen Nat Hist Soc. 25:3–16.
- Wei M, Liu Y, Guo H, Zhao F, Chen S. 2016. Characterization of the complete mitochondrial genome of Cynoglossus gracilis and a comparative analysis with other Cynoglossinae fishes. Gene. 591:369–375.
- Yokogawa K, Endo H, Sakaji H. 2008. Cynoglossus ochiaii, a new tongue sole from Japan (Pleuronectiformes: Cynoglossidae). Bull Natl Mus Nat Sci Ser A Suppl. 2:115–127.