Abstract
The complete chloroplast genome of Helixanthera parasitica from China was analyzed using next-generation sequencing. The plastome of H. parasitica is a typical quadripartite structure with a length of 125,021 bp, which contained inverted repeats (IR) of 22,752 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 73,151 bp and 6,366 bp, respectively. The cpDNA contains 105 genes, comprising 67 protein-coding genes, 30 tRNA genes, and 8 rRNA genes. The overall GC content of the plastome is 36.5%. Phylogenetic analysis showed that H. parasitica was closely related to the Tolypanthus maclurei.
Helixanthera parasitica Lour., belonging to family Loranthaceae, is mainly distributed in tropical countries such as Thailand, Laos, Indonesia, Cambodia, Vietnam, and Philippines, as well as in south and southwest China. Recorded hosts for this species include Castanopsis, Lithocarpus, Cinnamomum and Ficus sp. (Romina and Daniel Citation2008). Here, we report and characterize the complete plastome of H. parasitica based on Illumina paired-end sequencing data, which will contribute to the further studies on its genetic research and resource utilization. The annotated cp genome of H. parasitica has been deposited into GenBank with the accession number MN080718.
In this study, H. parasitica was sampled from in Fujian province of China, located at 117°21′32.85″ E, 24°29′24.24″ N. A voucher specimen (Y.-C. Shi et al. H065) was deposited in the Guangxi Key Laboratory of Plant Conservation and Restoration Ecology in Karst Terrain, Guangxi Institute of Botany, Guangxi Zhuang Autonomous Region and Chinese Academy of Sciences, Guilin, China. The experiment procedure is as reported in Zhang et al. (Citation2019). Around 2 Gb clean data were used for the cp genome de novo assembly by the program NOVOPlasty (Dierckxsens et al. Citation2017) and direct-viewing in Geneious R11 (Biomatters Ltd., Auckland, New Zealand). Annotation was performed with the program Plann (Huang and Cronk Citation2015) and Sequin (http://www.ncbi.nlm.nih.gov/).
The plastome of H. parasitica is a typical quadripartite structure with a length of 125,021 bp, which contained inverted repeats (IR) of 22,752 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 73,151 bp and 6,366 bp, respectively. The cpDNA contains 105 genes, comprising 67 protein-coding genes, 30 tRNA genes, and 8 rRNA genes. Among the annotated genes, 6 of them contain one intron (atpF, rpoC1, trnL-UAA, petB, petD and rpl2), and three genes (clpP, ycf3 and rps12) contain two introns. The overall GC content of the plastome is 36.5%, which is unevenly distributed across the whole chloroplast genome.
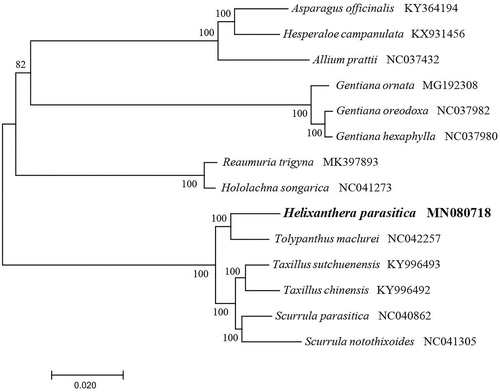
To identify the phylogenetic position of H. parasitica, phylogenetic analysis was conducted. A neighbor-joining (NJ) tree with 1000 bootstrap replicates was inferred using MEGA version 7 (Kumar et al. Citation2016) from alignments created by the MAFFT (Katoh and Standley Citation2013) using plastid genomes of 14 species. It showed the position of H. parasitica was closely related to the Tolypanthus maclurei (). Our findings will provide a foundation for facilitating its genetic research and contributing to its utilization in Loranthaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Romina VR, Daniel NL. 2008. Evolutionary relationships in the showy mistletoe family (Loranthaceae). Am J Bot. 95:1015–1029.
- Zhang Y, Shi YC, Duan N, Liu BB, Mi J. 2019. Complete chloroplast genome of Euphorbia tirucalli (Euphorbiaceae), a potential biofuel plant. Mitochondrial DNA B. 4:1973–1974.