Abstract
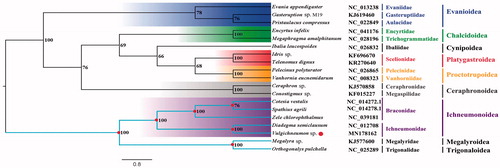
The Vulgichneumon sp. belongs to the subfamily Ichneumoninae of Ichneumonidae. The mitogenome (GenBank accession number: MN178162) of Vulgichneumon sp. was sequenced, the first representative of the mitogenome of the subfamily. The nearly complete mitogenome is 15,306 bp totally, consisting of 13 protein-coding genes, 2 rRNAs and 22 transfer RNAs. The nucleotide composition biases toward A and T, which together made up 83.6% of the entirety. Bayesian inference analysis supported the monophyly of Evanioidea, Chalcidoidea, Platygastroidea, Proctotrupoidea, Ceraphronoidea, and Ichneumonoidea. This result also suggested that the clade that contains other six superfamilies was the sister group to the clade that consist of Trigonaloidea, Megalyroidea and Ichneumonoidea.
Introduction
The Hymenoptera (ants, bees, and wasps) comprise one of the largest (LaSalle and Gauld Citation1993) and most biologically diverse group of insects (Gaston Citation1991) containing both eusocial and parasitic groups.
The specimens of Vulgichneumon sp. used for this study were collected from Weichang Manchu and Mongol Autonomous County of Heibei Province by Yimeng Chen and identified by Yimeng Chen. Specimens are deposited in the College of Plant Protection, Hainan University with the deposit number: HN NO.V20180926. The total genomic DNA was extracted from the whole body (except head) of the specimen using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) and stored at −20 °C until needed. The mitogenome was sequenced in Allwegene Technology Limited Company with the sequenced number: AWGT19042503-CC. The nearly complete mitogenome of Vulgichneumon sp. is 15,306 bp (GenBank accession number: MN178162). It encoded 13 PCGs, 22 tRNA genes, 2 rRNA genes and were similar with related reports before (Cha et al. Citation2007; Wei et al. Citation2009; Tan et al. Citation2011; Eimanifar et al. Citation2017). The nucleotide composition of the mitogenome was biased toward A and T, with 83.6% of A + T content (A = 41.2%, T = 42.4%, C = 10.4%, G = 6.0%). The A + T content of PCGs, tRNAs, and rRNAs is 82.0, 85.6, and 86.5%, respectively. The total length of all 13 PCGs of Vulgichneumon sp. is 10,966 bp. Six PCGs (NAD2, COI, ATP8, NAD3, NAD4, and NAD6) initiated with ATT codons and four PCGs (COIII, ATP6, NAD5, and CYTB) initiated with ATG codons, NAD1, NAD4L, and COII initiated with ATA as a start codon. Twelve PCGs used the typical termination codons TAA except NAD4 used T in Vulgichneumon sp.
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs from 19 Hymenoptera species. Bayesian (BI) analysis generated the phylogenetic tree topologies based on the PCGs matrices (). According to the phylogenetic result, the monophyly of Evanioidea, Chalcidoidea, Platygastroidea, Proctotrupoidea, Ceraphronoidea, and Ichneumonoidea were supported. The Megalyroidea was sister to Trigonaloidea and then Ichneumonoidea was assigned to the sister group to the clade of Megalyroidea + Trigonaloidea and the clade that contains other six superfamilies was the sister group to the clade that consists of Trigonaloidea, Megalyroidea, and Ichneumonoidea. This result that the phylogenetic relationship among Trigonaloidea, Megalyroidea, and Ichneumonoidea are consistent with the phylogenetic result of the previous study (Mao et al. Citation2015). The nearly complete mitogenome of Vulgichneumon sp. could provide important information for the further studies of Hymenoptera phylogeny.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, Jin BR, Han YS, Kim L. 2007. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene. 392:206–220.
- Eimanifar A, Kimball RT, Braun EL, Fuchs S, Grünewald B, Ellis JD. 2017. The complete mitochondrial genome of Apis mellifera meda (Insecta: Hymenoptera: Apidae). Mitochondr DNA B. 2:268–269.
- Gaston KJ. 1991. The magnitude of global insect species richness. Conserv Biol. 5:283–296.
- LaSalle J, Gauld ID. 1993. Hymenoptera and biodiversity. Wallingford, UK: CAB International.
- Mao M, Gibson T, Dowton M. 2015. Higher-level phylogeny of the Hymenoptera inferred from mitochondrial genomes. Mol Phylogenet Evol. 84:34–43.
- Tan H, Liu G, Dong X, Lin R, Song H, Huang S, Yuan Z, Zhao G, Zhu X. 2011. The complete mitochondrial genome of the Asiatic cavity-nesting honeybee Apis cerana (Hymenoptera: Apidae). PLoS One. 6:e23008.
- Wei S, Shi M, He J, Sharkey M, Chen X. 2009. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome. 52:308–319.