Abstract
Ceutorhynchus obstrictus (Marsham, 1802) is a serious pest of oilseed rape (Brassica napus L.) in Europe and the USA. We have determined a 20,124 bp mitogenome of C. obstrictus which includes 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 2,773 bp. The base composition was AT-biased (81.4%). Hypothetical ORFs are identified in the control region. Phylogenetic trees present that C. oibstricus is clustered with Alcides juglans (Alcidinae). It also shows polyphyletic manner for two tribes, requiring more mitogenomes to resolve it.
Ceutorhynchus obstrictus (Marsham, 1802), also known as cabbage seedpod weevil, is a major pest of oilseed rape (Brassica napus) originated in Europe. It has successfully settled across the globe including the U.S. (Cárcamo et al. Citation2001). It poses a major threat to economic sustainability of canola production in western Canada (Cárcamo et al. Citation2001). In 1995, C. obstrictus was first collected from Gimhae region, Korea, consequently became a serious insect pest on B. napus in Korea (Kim et al. Citation2018). To understand its genetic background, we determined its complete mitogenome as first mitogenome in Ceutorhynchinae.
Genomic DNA of C. obstrictus collected from Seogwipo-si, Jeju-do in Korea in 2019 (33°52′50ʺN, 126°93′04ʺE; specimen is stored in Gyeongsang National University, Korea, Accession number: Coll#HB002) was extracted using DNeasy Brood & Tissue Kit (QIAGEN, Hilden, Germany). HiSeqX was used for sequencing (Macrogen Inc., Seoul, Korea). Filtering, de novo assembly, and gap-filling processes were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), Trimmomatic 0.33 (Bolger et al. Citation2014), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAM tools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) and ARWEN (Laslett and Canbäck Citation2008) were used to annotate mitogenome of C. obstrictus based on Eucryptorhynchus brandti mitogenome (Nan et al. Citation2016).
Cryptopone obstrictus mitogenome (MN180050) is 20,124 bp and GC ratio is 18.6%. It contains 13 protein-coding genes (PCGs), 2 rRNAs, and 22 tRNAs. Range of tRNA size is 64–71 bp, which is smaller than those of some insect species, such as Aiolocaria hexaspilota (55–70 bp; Seo et al. Citation2019) and Cryptopone sauteri (56–78 bp; Park, Kwon, Park, Citation2019). Gene order of C. obstrictus is similar to mitogenomes of other weevils, which is the ancestral gene order of all insects. Interestingly, inside the control region, there are two hypothetical ORFs of which directions are reversed in the same coordination and amino acids are different. No homologous genes of them are found in non-redundant database. Mitogenome of Scolytinae sp. (KX035192) also shows the same phenomenon. In addition, mitogenomes of Anisandrus dispar (NC_036293), Curculionidae sp. (KX035176), and Hypothenemus sp. (KX035163) also have a hypothetical ORF in control region, indicating not exceptional case. It may be explained by expansion of fungal mitogenomes of genus Aspergillus (Xu et al. Citation2018; Park et al. Citation2019).
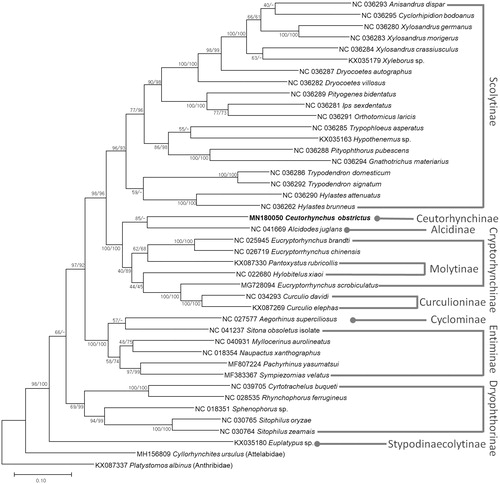
We inferred the phylogenetic relationship of 38 Curculionid species including C. obstrictus and two outgroup species of Cyllorhynchites ursulus (MH156809) and Platystomos albinus (KX087337). Multiple sequence alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013) using concatenated alignments of all PCGs. Bootstrapped maximum likelihood and neighbor-joining trees were constructed using MEGA X (Kumar et al. Citation2018). Ceutorhynchus obstrictus (Ceutorhynchinae) was clustered with Alcides juglans (Alcidinae) with relatively low bootstrap value in both trees (), indicating that more mitogenomes in both subfamilies are required to unravel their phylogenetic relationship. Phylogenetic trees present paraphyletic manners in two subfamilies, Cryptorhynchinae and Entiminae, with relative low bootstrap values (). Our mitogenome will provide phylogenetic insights in Curculionidae family in near future.
Figure 1. Maximum likelihood (bootstrap repeat is 1000)and neighbor-joining (bootstrap repeat is 10,000) phylogenetic tree of 38 Curculionidae species: Ceutorhynchus obstrictus (MN180050: this study), Anisandrus dispar (NC 036293), Cyclorhipidion bodoanus (NC 036295), Xylosandrus germanus (NC 036280), Xylosandrus morigerus (NC 036283), Xylosandrus crassiusculus (NC 036284), Xyleborus sp. (KX035179), Dryocoetes autographus (NC 036287), Dryocoetes villosus (NC 036282), Pityogenes bidentatus (NC 036289), Ips sexdentatus (NC 036281), Orthotomicus laricis (NC 036291), Trypophloeus asperatus (NC 036285), Hypothenemus sp. (KX035163), Pityophthorus pubescens (NC 036288), Gnathotrichus materiarius (NC 036294), Trypodendron domesticum (NC 036286), Trypodendron signatum (NC 036292), Hylastes attenuatus (NC 036290), Hylastes brunneus (NC 036262), Alcidodes juglans (NC 041669), Eucryptorhynchus brandti (NC 025945), Eucryptorrhynchus chinensis (NC 026719), Pantoxystus rubricollis (KX087330), Hylobitelus xiaoi (NC 022680), Eucryptorrhynchus scrobiculatus (MG728094), Curculio davidi (NC 034293), Curculio elephas (KX087269), Aegorhinus superciliosus (NC 027577), Sitona obsoletus (NC 041237), Myllocerinus aurolineatus (NC 040931), Naupactus xanthographus (NC 018354), Pachyrhinus yasumatsui (MF807224), Sympiezomias velatus (MF383367), Cyrtotrachelus buqueti (NC 039705), Rhynchophorus ferrugineus (NC 028535), Sphenophorus sp. (NC 018351), Sitophilus oryzae (NC 030765), Sitophilus zeamais (NC 030764), Euplatypus sp. (KX035180), and two outgroup species: Cyllorhynchites ursulus (MH156809, Attelabidae), Platystomos albinus (KX087337, Anthribidae). Phylogenetic tree was drawn based on maximum likelihood phylogenetic tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Cárcamo H, Dosdall L, Dolinski M, Olfert O, Byers J. 2001. The cabbage seedpod weevil, Ceutorhynchus obstrictus (Coleoptera: Curculionidae)-a review. J Entomol Soc British Columbia. 98:201–210.
- Kim K, Lee W, Hong KJ. 2018. Ecological replacement of native rapeseed weevil (Ceutorhynchus albosuturalis) by invasive alien species, cabbage seedpod weevil (C. obstrictus) on rapeseed flowers in Korea. Korean J Appl Entomol. 57:323–328.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:13033997. Preprint at https://arxiv.org/abs/1303.3997
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Nan X, Wei C, He H. 2016. The complete mitogenome of Eucryptorrhynchus brandti (Harold)(Insecta: Coleoptera: Curculionidae). Mitochondr DNA A. 27:2060–2061.
- Park J, Kwon W, Huang X, Mageswari A, Heo I-B, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of a xerophilic fungus, Aspergillus pseudoglaucus. Mitochondr DNA B. 4:2422–2423.
- Park J, Kwon W, Park J. 2019. The complete mitochondrial genome of Cryptopone sauteri Wheeler, WM, 1906 (Hymenoptera: Formicidae). Mitochondr DNA B. 4:614–615.
- Seo BY, Park J, Kwon W, Park J. 2019. The complete mitochondrial genome of Aiolocaria hexaspilota (Hope, 1831)(Coleoptera: Coccinellidae). Mitochondr DNA B. 4:1472–1474.
- Xu Z, Wu L, Liu S, Chen Y, Zhao Y, Yang G. 2018. Structure characteristics of Aspergillus egyptiacus mitochondrial genome, an important fungus during the fermentation of dark tea. Mitochondr DNA B. 3:1135–1136.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
