Abstract
In Japan, two closely-related damselflies, Mnais costalis Selys, 1869 (Odonata: Calopterygidae) and M. pruinosa Selys-Longchamps (Odonata: Calopterygidae), 1853, coexist, and they exhibit geographic variations in wing color, body size, and habitat preference. In this study, we analyzed the complete mitochondrial genome of M. costalis from Saga Prefecture, Japan (sympatric populations that exhibit wing color polymorphism), and compared the genome with M. costalis that exhibit monomorphic orange wing color. The mitochondrial genome of M. costalis from Saga Prefecture was identified as a circular molecule of 15,488 bp, similar to that found in other M. costalis populations. It was predicted to contain 13 protein-coding (PCG), 22 tRNA, and two rRNA genes, along with one A + T-rich control region. Among the PCGs, ATP8 and ATP6, ATP6 and COIII, ND4 and ND4L, and ND6 and Cytb shared seven, one, seven, and one nucleotides, respectively. The initiation codon ATG was found in eight genes, ATC in four, and ATT in one, while the termination codons TAA, TAG, TA, and T were observed in seven, one, two, and three genes, respectively. All the tRNA genes possessed a cloverleaf secondary structure, except for tRNA-His that lacks the TΨC loop. The average AT content of mitochondrial genome was 66.06%. From a phylogenetic analysis, the loss of wing color polymorphism in monomorphic sympatric populations is likely to occur with the coexistence of two Mnais species.
Mnais costalis Selys, 1869 (Odonata: Calopterygidae) often coexists with the closely related damselfly, M. pruinosa Selys 1853. These species exhibit geographic variations of wing color, body size, and habitat preference (e.g. Suzuki and Tamaishi Citation1982; Nomakuchi et al. Citation1984; Siva-Jothy and Tsubaki Citation1989; Okuyama et al. Citation2013; Tsubaki and Okuyama Citation2016). In allopatric populations, M. costalis males exhibit wing color polymorphism (Tsubaki et al. Citation1997), with orange morphs and clear morphs being observed. In sympatric populations of two Mnais species in the Chugoku, Shikoku, and Kyusyu regions of Japan, M. costalis males exhibit the same wing color polymorphism as allopatric populations, but are monomorphically orange-winged in the Chubu and Kinki regions. In this study, we analyzed the complete mitochondrial genome of M. costalis from a sympatric population in Kyusyu, and compared the genome with M. costalis from a sympatric population in Kinki (AP017642; Okuyama and Takahashi Citation2017), and from an allopatric population (KU871065; Lorenzo-Carballa et al. Citation2016).
We collected adult orange-winged male M. costalis individuals from Saga Prefecture, Japan in May 2017. The adult damselflies were transferred immediately to 99.5% ethanol. The specimen was stored the National Museum of Nature and Science, Japan (accession number: NSMT-I-Od-33337). Genomic DNA isolated from one damselfly was sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). The complete mitochondrial genome of M. costalis (AP017642) was used as a reference sequence. The resultant reads were assembled and annotated using the MITOS web server (Bernt et al. Citation2013) and Geneious R9 (Biomatters). These 13 protein-coding genes (PCGs) and two rRNA genes sequences were aligned using MEGA6 (Tamura et al. Citation2013). The phylogenetic analysis was performed with the maximum likelihood (ML) criterion using TREEFINDER (Jobb Citation2011).
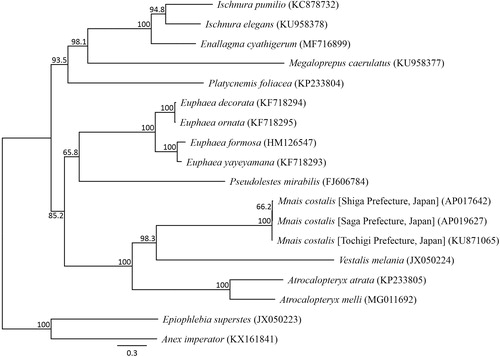
We succeeded in sequencing the entire mitochondrial genome of M. costalis from Saga Prefecture (DDBJ accession number AP019627). The genome consisted of a closed loop 15,488 bp-long, which included 13 PCGs, 22 tRNA genes, two rRNA genes, and one AT-rich control region, which resembles the genomic organization common in M. costalis. The heavy strand was predicted to have nine protein-coding and 14 tRNA genes, while the light strand was predicted to contain four protein-coding, eight tRNA, and two rRNA genes. Eight PCGs started with ATG; the ND2, ATP8, ND3, and ND6 genes with ATC; and the ND5 gene with ATT. Seven PCGs used the stop codon TAA; the ND5 gene used TAG; the COIII and ND4 genes used the incomplete stop codon TA; and the COII, ND5, and Cytb genes used a single T. All the tRNA genes possessed a cloverleaf secondary structure, except for tRNA-His. A phylogenetic analysis was constructed using 13 PCGs across 14 Zygoptera taxa (). The phylogenetic relationships of three M. costalis populations suggested that the loss of wing color polymorphism in the Chubu and Kinki regions may have occurred after coexistence with M. pruinosa. However, this relationship is supported with a lower bootstrap value, therefore it is necessary to undertake further analysis of M. costalis mitochondrial genome data from these three distinct populations.
Figure 1. Phylogenetic relationships (maximum likelihood) of the Zygoptera based on the nucleotide sequences of the 13 protein-coding genes of the mitochondrial genome. Sequences from Epiophlebia superstes (JX050223, Wang et al. Citation2015) and Anax imperator (KX161841, Feindt et al. Citation2016) were used as an outgroup. These sequences were separated by codon positions, and for each partition, the optimal models of sequence evolution were used in the maximum likelihood method using TREEFINDER, based on the corrected Akaike information criterion. The numbers at the nodes indicate the bootstrap support inferred from 1000 bootstrap replicates. Alphanumeric terms indicate the DNA Database of Japan accession numbers.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Feindt W, Osigus HJ, Herzog R, Mason CE, Hadrys H. 2016. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA Part B. 1:497–499.
- Jobb G. 2011. TREEFINDER version of March 2011. Munich. http://www.treefnder.de.
- Lorenzo-Carballa MO, Tsubaki Y, Plaistow SJ, Phillip C. 2016. The complete mitochondrial genome of the broad-winged damselfly Mnais costalis Selys (Odonata: Calopterygidae) obtained by next-generation sequencing. Int J Odonat. 19:91–98.
- Nomakuchi S, Higashi K, Harada M, Maeda M. 1984. An experimental study of the territoriality in Mnais pruinosa pruinosa Selys (Zygoptera: Calopterygidae). Odonatologica. 13:259–267.
- Okuyama H, Samejima Y, Tsubaki Y. 2013. Habitat segregation of sympatric Mnais damselflies (Odonata: Calopterygidae): microhabitat insolation preferences and competition for territorial space. Int J Odonat. 16:109–117.
- Okuyama H, Takahashi J. 2017. The complete mitochondrial genome of Mnais costalis Selys (Odonata: Calopterygidae) assembled from next generation sequencing data. TOMBO. 59:77–83.
- Siva-Jothy MT, Tsubaki Y. 1989. Variation in copulation duration in Mnais pruinosa pruinosa Selys (Odonata: Calopterygidae). 1. Alternative mate-securing tactics and sperm precedence. Behav Ecol Sociobiol. 25:261–267.
- Suzuki K, Tamaishi A. 1982. Ethological study on two Mnais species, Mnais nawai Yamamoto and M. pruinosa Selys, in the Hokuriku District, central Honshu, Japan. Analysis of adult behavior by marking-reobservation experiments. J Coll Liberal Arts Toyama Univ (Nat Sci). 14:95–128.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tsubaki Y, Hooper RE, Siva-Jothy MT. 1997. Differences in adult and reproductive lifespan in the two male forms of Mnais pruinosa costalis selys (Odonata: Calopterygidae). Res Popul Ecol. 39:149–155.
- Tsubaki Y, Okuyama H. 2016. Adaptive loss of color polymorphism and character displacements in sympatric Mnais damselflies. Evol Ecol. 30:1–14.
- Wang JF, Chen MY, Chaw SM, Morii Y, Yoshimura M, Sota T, Lin CP. 2015. Complete mitochondrial genome of an enigmatic dragonfly, Epiophlebia superstes (Odonata, Epiophlebiidae). Mitochondrial DNA. 26:718–719.
