Abstract
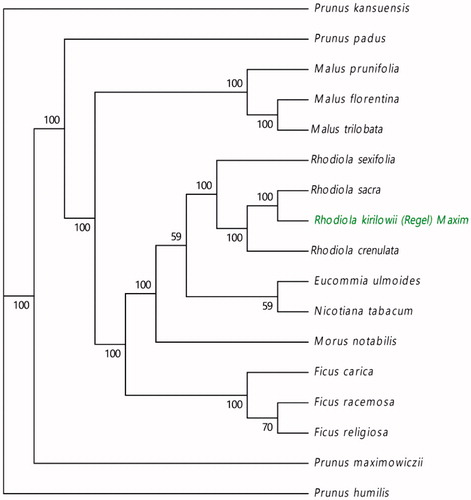
Rhodiola kirilowii is a precious Tibetan drug and an extremely endangered plant. In recent years, the number of individuals of R. kirilowii has decreased sharply. Here, we determined and analyzed the complete chloroplast genome of R. kirilowii. The cpDNA was 150,905 bp in length, containing a pair of inverted repeats (IRs) of 25,864 bp each separated by a large and small single copy (LSC and SSC) regions of 82,131 bp and 17,046 bp, respectively. The genome contained 84 protein-coding genes, 8 rRNA genes, and 36 tRNA genes. The overall GC content of the chloroplast genome was 37.8%, whereas the corresponding values of the LSC, SSC, and IR regions were 35.8, 31.7 and 42.9%, respectively. A maximum likelihood (ML) phylogenetic analysis demonstrated that Rhodiola sacra and Rhodiola crenulata were clustered into one clade with strong support values, indicating their closer relationship.
Rhodiola kirilowii was a rare traditional Chinese drug resource. Modern medicine proves that extracts from R. kirilowii contain a variety of chemical constituents, and have various pharmacological effects such as anti-hypoxia, fatigue, tumor, radiation, aging, and improvement of mental and physical functions (Hong et al. Citation2017). Rhodiola kirilowii usually grows in slabs under harsh and variable environmental conditions such as alpine tundra and hillside forests (Liu et al. Citation2015). Coupled with unrestricted collection, the number of wild resources has dropped dramatically. Therefore, it is extremely urgent to survey the genetic background so as to facilitate the preservation of R. kirilowii. The present study is the first time to assemble and characterize the complete chloroplast genome for R. kirilowii (GenBank: MN109979) using the Illumina pair-end sequencing reads. We believe that such information will provide the underlying information for genetic and conservation studies and explore possible phylogenetic relationship within Crassulaceae family.
The plant materials of R. kirilowii were collected from Nyingchi (Tibet, China; N:29°13ʹ08.12”, E:092°41ʹ51.83”). The specimens of R. kirilowii have been kept in Tibet Agriculture & Animal Husbandry University. Total genomic DNA of R. kirilowii was extracted from silica-gel-dried leaves using the modified CTAB method (Allen et al. Citation2006). Sequencing was carried out on the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA). A total of 2.3 Gb raw reads were obtained and then de novo assembled using CLC genome assembler program (ver.4.06 beta, CLC Inc, Aarhus, Denmark) as previously described (Kim et al. Citation2015). We assembled the chloroplast genome using Geneious Prime 2019.2.1(Kearse et al. Citation2012), with Sedum oryzifolium (GenBank: NC027837) as the reference. DOGMA was used for annotation of the complete chloroplast genome (Wyman et al. Citation2004), and the annotation was corrected with Geneious Prime 2019.2.1 (Kearse et al. Citation2012).
The complete cp genome of R. kirilowii is 150,905 bp in length and encodes 128 genes, including 84 protein-coding genes, 36 transfer RNA genes, and 8 ribosomal RNA genes. Its quadripartite structure was composed of two inverted repeat (IRa and IRb) regions of 25,864 bp, a large single copy (LSC) region of 82,131 bp and a small single copy (SSC) region of 17,046 bp. Among annotated genes, 14 genes (trnK-UUU, rps16, atpF, rpoC1, trnV-UAC, trnL-UAA, petB, petD, rpl16, rpl2, ndhB, ndhA, trnA-UGC and trnI-GAU,) harbored one intron and three genes (ycf3, clpP and rps12) harbored two introns. The base composition of R. kirilowii cp genome was uneven (30.8% A, 19.2% C, 18.5% G, 31.4% T) with an overall GC content of 37.8% and the corresponding values of the LSC, SSC and IR regions reaching 35.8, 31.7 and 42.9%, respectively.
In order to ascertain the phylogenetic position of R. kirilowii, a maximum likelihood (ML) phylogenetic tree was constructed with MEGA7(Kumar et al. Citation2016). A maximum likelihood (ML) phylogenetic () analysis demonstrated that Rhodiola sacra and Rhodiola crenulate were clustered into one clade with strong support values, indicating their closer relationship. The complete cp genome information reported in this paper provided data useful for both populations genomic studies and conservation works on R. kirilowii. This genome will also contribute significantly to the phylogenetic and evolutionary studies of the genus Rhodiola L.
Figure 1. Phylogenetic tree based on the complete chloroplast genome sequences of R. sacra and 16 other species. The tree was generated using a ML method by MEGA7 with 1000 bootstrap replicates. Numbers on the nodes indicate bootstrap values. The chloroplast genome sequences used to construct the phylogenetic tree are MF766010, KY635880, KT368151, KY416513, KX499856, KU851961, KX499858, MK301435, MF405921, KF990036, KP760071, KP760072, Z00044, MN218690, MN109979, MN109978, MN109980.
Disclosure statement
The authors report no declaration of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320.
- Hong DX, Su JS, Wen J, Zhang J, Zhang Y. 2017. [Resource investigation about Tibetan medicine Rhodiola kirilowii]. Zhongguo Zhong Yao Za Zhi. 42(6):1202–1206.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870.
- Liu P, Liu J, Zhang G, Zhang X, Zhu H, Youfang LI. 2015. Study on Breaking Resistance Characteristics of Four Kinds of Plants' Lateral Root Branch During the Vigorous Growth Period. Northern Horticulture.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
