Abstract
Fusarium oxysporum is a famous plant pathogenic filamentous fungus. Here, we report the complete mitochondrial genome sequence of F. oxysporum f. sp. lactucae isolated from the lettuce field in Suwon area, Korea. Total length of the mitochondrial genome is 45,020 bp and it encodes 42 genes (15 protein-coding genes, two rRNAs, and 25 tRNAs). Nucleotide sequence of coding region takes over 32.7%, and overall GC content is 32.4%. Phylogenetic tree presented that F. oxysporum f. sp. lactucae 09-002 was clustered with Fusarium commune not like another F. oxysporum mitochondrial genomes, requiring further analyses.
Fusarium oxysporum is an ascomycete and soilborne fungus in every type of soils all over world (Alabouvette and Couteaudier Citation1992). It is a famous plant pathogenic filamentous fungus which can infect many plant species including crops (Fravel et al. Citation2003). More than 120 formae speciales and races have been classified in F. oxysporum (Armstrong Citation1981). At least 62 mitogenome sequences of F. oxysporum have been completed and compared (Pantou et al. Citation2008; Fourie et al. Citation2013; Brankovics et al. Citation2017), presenting diverse features of its mitochondrial genomes as other fungal mitogenomes are (Joardar et al. Citation2012; Xu et al. 2018; Chen et al. Citation2019; Park, Kwon, Huang, et al. Citation2019; Park, Kwon, Zhu, Mageswari, Heo, Han, et al. Citation2019; Park, Kwon, Zhu, Mageswari, Heo, Kim, et al. Citation2019). Fusarium oxysporum f. sp. lactucae 09-002 isolated from wilted lettuce in Suwon area, South Korea (37.28036 N, 127.00870E), was identified based on translation elongation factor 1a gene (Samson et al. Citation2004). Here, we completed its mitochondrial genome as a first mitogenome in F. oxysporum f. sp. lactuace.
The hyphae of F. oxysporum were collected from samples taken from Horticultural and Herbal Crop Environment Division (F. oxysporum, f. sp. lactucae 09-002) and its DNA was extracted by using a HiGene™ Genomic DNA Prep Kit (BIOFACT, Korea). Raw data generated by HiSeq4000 were subject to de novo assembly done by Velvet 1.2.10 (Zerbino and Birney 2008), gap filling with SOAPGapCloser 1.12 (Zhao et al. 2011), and base confirmation with BWA 0.7.17 and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate its mitogenome by comparing with that of Fusarium commune strain JCM11502 (NC_036106; Brankovics et al. Citation2017).
Length of F. oxysporum f. sp. lactucae 09-002 mitogenome (Genbank accession is MN259515) is 45,020 bp, which is eighth shortest mitogenome among sixty-four mitogenomes (from 34,477 bp to 53,639 bp). It encodes 42 genes consisting of 15 protein-coding genes (PCGs), two rRNAs, and 25 tRNAs, which is similar to those of another F. oxysporum. Nucleotide sequence of coding region takes over 32.7%. Overall GC content of this mitochondrial genome is 32.4%, which is the highest GC ratio among 62 mitochondrial genomes of F. oxysporium.
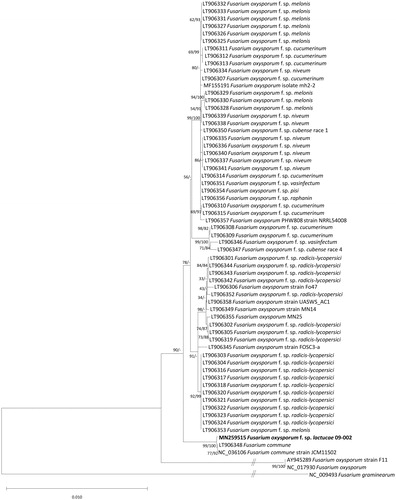
Sequence alignment of conserved PCGs extracted from sixty-two F. oxysporum mitogenomes, two F. commune, and one F. graminearum (Al-Reedy et al. Citation2012) as an outgroup was conducted by MAFFT 7.388 (Katoh and Standley Citation2013). The neighbor joining and maximum likelihood phylogenetic trees were constructed using MEGA X (Kumar et al. Citation2018) with 10,000 and 1,000 bootstrap repeats, respectively. Phylogenetic tree presents that our mitogenome is clearly clustered with two mitogenome of F.commune (). In addition, elongation factor 1a sequence of both species are similar to each other, causing confusion of species identification. Since our strain was isolated from the wilted lettuce, we made a conclusion that it was F. oxysporium rather than F. commune, which would be confirmed later using more available mitochondrial genomes of F. oxysporium and F. commune in the near future.
Figure 1. Neighbor joining and maximumlikelihood phylogenetic trees (bootstrap repeat is 10,000 and 1,000, respectively) of 64 Fusarium mitochondrial genomes: F. oxysporum f. sp. lactucae (MN259515 in this study), F. oxysporum (NC_017930, AY874423, AY945289, MF155191, LT906306, LT906349, LT906355, LT906358, LT906357, and LT906345), F. oxysporum f. sp. melonis (LT906328, LT906329, LT906330, LT906325, LT906326, LT906327, LT906331, LT906332, LT906333, and LT906353), F. oxysporum f. sp. pisi (LT906354), F. oxysporum f. sp. vasinfectum (LT906351), F. oxysporum f. sp. raphanin (LT906356), F. oxysporum f. sp. cubense race 1 (LT906350), F. oxysporum f. sp. cubense race 4 (LT906347), F. oxysporum f. sp. niveum (LT906334, LT906338, LT906335, LT906336, LT906340, LT906341, LT906339, and LT906337), F. oxysporum f. sp. cucumerinum (LT906315, LT906314, LT906310, LT906307, LT906308, LT906309, LT906311, LT906312, and LT906313), F. oxysporum f. sp. radicis-lycopersici (LT906352, LT906342, LT906343, LT906344, LT906302, LT906305, LT906319, LT906301, LT906303, LT906304, LT906316, LT906317, LT906318, LT906320, LT906321, LT906322, LT906323, and LT906324), F. oxysporum f. sp. vasinfectum (LT906346), F. commune (LT906348 and NC_036106), and F. graminearum (NC_009493) as an outgroup. Phylogenetic tree was drawn based on neighborjoining tree. Numbers on branches indicate bootstrap values of neighbor joining and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Al-Reedy RM, Malireddy R, Dillman CB, Kennell JC. 2012. Comparative analysis of Fusarium mitochondrial genomes reveals a highly variable region that encodes an exceptionally large open reading frame. Fungal Genet Biol. 49:2–14.
- Alabouvette C, Couteaudier Y. 1992. Biological control of Fusarium wilts with nonpathogenic Fusaria. Biological control of plant diseases. Springer; p. 415–426.
- Armstrong G. 1981. Formae speciales and races of Fusarium oxysporum causing wilt disease. Fusarium: Disease, Biology, and Taxonomy.:391–399. University Park, PA, USA: Pennsylvania State University Press.
- Brankovics B, van Dam P, Rep M, de Hoog GS, van der Lee TA, Waalwijk C, van Diepeningen AD. 2017. Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex. BMC Genomics. 18:735.
- Chen C, Fu R, Wang J, Hu R, Li X, Luo X, Chen X, Lu D. 2019. Characterization and phylogenetic analysis of the complete mitochondrial genome of Aspergillus sp. (Eurotiales: Eurotiomycetidae). Mitochondrial DNA B. 4:752–753.
- Fourie G, Van der Merwe NA, Wingfield BD, Bogale M, Tudzynski B, Wingfield MJ, Steenkamp ET. 2013. Evidence for inter-specific recombination among the mitochondrial genomes of Fusarium species in the Gibberella fujikuroi complex. BMC Genomics. 14:605.
- Fravel D, Olivain C, Alabouvette C. 2003. Fusarium oxysporum and its biocontrol. New Phytol. 157:493–502.
- Joardar V, Abrams NF, Hostetler J, Paukstelis PJ, Pakala S, Pakala SB, Zafar N, Abolude OO, Payne G, Andrianopoulos A, et al. 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics. 13:698.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:13033997. Utrecht: Centraalbureau voor Schimmelcultures.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Pantou MP, Kouvelis VN, Typas MA. 2008. The complete mitochondrial genome of Fusarium oxysporum: insights into fungal mitochondrial evolution. Gene. 419:7–15.
- Park J, Kwon W, Huang X, Mageswari A, Heo I-B, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of a xerophilic fungus, Aspergillus pseudoglaucus. Mitochondrial DNA B. 4:2422–2423.
- Park J, Kwon W, Zhu B, Mageswari A, Heo I-B, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of the food fermentation fungus, Aspergillus luchuensis. Mitochondrial DNA B. 4:945–946.
- Park J, Kwon W, Zhu B, Mageswari A, Heo I-B, Kim J-H, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of an aflatoxin B and G producing fungus, Aspergillus parasiticus. Mitochondrial DNA B. 4:947–948.
- Samson RA, Hoekstra ES, Frisvad JC. 2004. Introduction to food-and airborne fungi. In: Tjamos E.C., Papavizas G.C., Cook R.J., editors. Centraalbureau voor Schimmelcultures (CBS). 7th ed. Boston, MA.
