Abstract
Caulerpa lentillifera is a marine nutrient-rich edible green algae, with its external shape similar to ‘sea grape’, it has functions of purifying blood, anti-oxidation, anti-cancer, and anti-tumor. The mitogenome sequence of C. lentillifera is 209,894 bp long. A total of 67 genes were determined, including 17 protein-encoding genes, 3 rRNA genes, 27 tRNA genes, and 20 unidentified open reading frame (ORF). Phylogenetic analysis showed that C. lentillifera clustered together into a single branch. The mitogenome analysis will help the understanding of Ulvophyceae evolution.
Caulerpa lentillifera is a kind of nutrient-rich edible green algae and it contains a variety of amino acids and vitamins needed by the human body (Guo et al. Citation2015). It is also known as ‘green caviar’ in Japan and is widely distributed in the tropical waters of the tropical edible economic algae. Caulerpa lentillifera belongs to Chlorophyta, Ulvophyceae, Bryopsodales, Caulerpaceae, Caulerpa. It is also effective in treating hyperglycemia, anti-oxidation, and enhancing immunity (Maeda et al. Citation2012; Paul et al. Citation2014). In this study, we sequenced, assembled, and annotated the mitochondrial DNA of C. lentillifera, conducted a systematic genome study to evaluate the evolutionary trend of mitochondrial DNA, compared with others submitted in NCBI, and studied the phylogenetic relationship between C. lentillifera and other green algae.
The determination of the complete C. lentillifera mitogenome sequence by next-generation sequencing methods was conducted. The specimen was collected from north China (Qionghai, Hainan Province, 19°14′36.06″N,110°28′13.01″E) and stored at the Culture Collection of Seaweed at the Ocean University of China (accession number: 2018050001). Total DNA was extracted using the modified CTAB method (Doyle and Doyle Citation1990). Paired-end reads (150 bp) were sequenced by using Illumina HiSeq system (Illumina, San Diego, CA) and Pacbio system. tRNAscan-SE Search Server (Schattner et al. Citation2005) was used to identify the tRNA genes. The other mitogenomic regions were annotated with Geneious R10 (Biomatters Ltd, Auckland, New Zealand), using the C. lentillifera (NC_038217) mitogenome as reference.
The complete C. lentillifera mitogenome is a circular DNA molecule measuring 209,894 bp in length with the overall G + C content of 50.9% (GenBank accession number:MN201586). The mitogenome contains 67 genes, including 17 protein-coding, 3 rRNA, 27 tRNA genes, and 20 unidentified open reading frame (ORF). The nucleotide composition was 24.92% A, 25.81% C, 25.08% G, and 24.19% T. The length of the coding region was 106,258 bp, corresponding to 50.62% of the total length. Of the 17 protein-coding genes, 9 (52.94%) ended with the TAA stop codon, 4 (23.53%) with TAG (cox1, cytb, ND2, and ND6) and 4 (23.53%) with TGA(atp6, atp9, cox3, and ND4). All protein-coding genes in C. lentillifera were found to use the start codon ATG. The lengths of 3 rRNA genes are 2214 bp (rrn23 rRNA), 110 bp (rrn5 rRNA), and 1454 bp (rrn16 rRNA). The gene numbers and structures were largely similar to C. lentillifera published in the NCBI database, but it is 860 bp longer than the reference, it was verified by amplification experiment that the 860 bp was indeed present in the mitochondrial genome of C. lentillifera, which is correct.
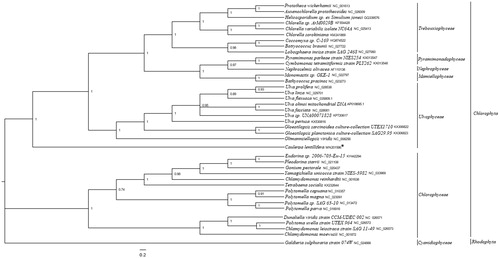
Five shared mitochondrion protein sequences from 39 green algae and 1 outgroup including C. lentillifera were used to conduct phylogenetic analysis by using MrBayes 3.1.2 software (Ronquist and Huelsenbeck Citation2003). Galdieria sulphuraria strain 074W(NC_024666) served as the outgroup. Poorly aligned regions were removed by using the Gblocks server (Castresana Citation2000). Caulerpa lentillifera clustered together into a single branch and showed a further relationship with Ulvophyceae species (). This complete mitogenome analysis of C. lentillifera helps us better understand the evolutionary process of Ulvophyceae.
Disclosure statement
No conflict of interest, for all authors, including the implementation of research experiments and writing of this article was reported.
Additional information
Funding
Reference
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- Guo H, Yao J, Sun Z, Duan D. 2015. Effects of salinity and nutrients on the growth and chlorophyll fluorescence of Caulerpa lentillifera. Chin J Ocean Limnol. 33:410–418.
- Maeda R, Ida T, Ihara H, et al. 2012. Induction of apoptosis in MCF-7 cells by β-1,3-xylooligosaccharides prepared from Caulerpa lentillifera. Biosci Biotechnol Biochem. 76:1032–1034.
- Paul NA, Neveux N, Magnusson M, et al. 2014. Comparative production and nutritional value of “sea grapes” — the tropical green seaweeds Caulerpa lentillifera and C. racemosa. J Appl Phycol. 26:1833–1844.
- Ronquist F, Huelsenbeck JP. 2003. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.