Abstract
The complete mitochondrial genome of Pisaura bicornis (GenBank accession number MN296112) is 15,281 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes, and a putative control region. The overall base composition of this genome is A (35.65%), C (8.12%), G (13.47%) and T (42.76%), demonstrating an obvious bias of high AT content (73.2%). ATT, ATA, TTG were initiation codons and TAA, TAG, and T were termination codons. Ten tRNAs (trnD, trnF, trnG, trnH, trnK, trnP, trnR, trnT, trnY, and trnE) lacked the TΨC arm stem, while three tRNAs (trnA, trnS1, and trnS2) lost the dihydrouracil (DHU) arm. Phylogenetic tree based on 13 PCGs showed that P. bicornis is closely related to Dolomedes angustivirgatus and belongs to Pisauridae.
Keywords:
The nursery-web spider Pisaura bicornis belongs to the family Pisauridae, which comprises more than 300 species in 48 genera around the world (Sierwald Citation1997; Platnick Citation2015). Most of these spiders are carnivores and often live in farmland or near streams (Zhang Citation2000). In this study, adult species of P. bicornis were collected from Maolan Nature Reserve in Libo county (N25°20′, E107°55′), Guizhou Province, China, and deposited in the spider specimen room of Guiyang University with an accession number GYU-GZML-06.
The complete mitogenome of P. bicornis (GenBank accession number MN296112) is 15,281 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and a putative control region. The gene content and arrangement of P. bicornis are similar to those found in previously determined spider mitogenomes (Li et al. Citation2016; Yang et al. Citation2019). Four PCGs (nad5, nad4, nad4L, and nad1), nine tRNAs (trnY, trnC, trnL2, trnF, trnH, trnP, trnL1, trnV, and trnQ), and two rRNAs (rrnL and rrnS) were encoded on the light strand (N-strand). Nine PCGs (nad2, cox1, cox2, cox3, atp6, atp8, nad3, nad6, and cytb) and thirteen tRNAs (trnM, trnW, trnK, trnD, trnG, trnN, trnA, trnS1, trnR, trnE, trnI, trnS2, and trnT) were encoded on the heavy strand (J-strand). The overall base composition of P. bicornis mitogenome was A (35.65%), C (8.12%), G (13.47%) and T (42.76%), with a high AT bias of 73.2%. The AT-skew and GC-skew of this mitogenome were −0.091 and 0.248, respectively.
Gene overlaps were found in 23 locations and involved a total of 214 bp. The longest overlap is 45 bp in length and located between nad4 and nad4L. There are 8 intergenic spacer regions comprising a total of length of 49 bp and the largest spacer (15 bp) resided between trnP and nad6. The length of 22 tRNAs ranged from 48 bp (trnK) to 70 bp (trnI), The A + T content ranged from 69.09% (trnR) to 88.14% (trnC). Thirteen tRNAs lacked the potential to form the cloverleaf secondary structure. Ten of them (trnD, trnF, trnG, trnH, trnK, trnP, trnR, trnT, trnY, and trnE) lacked the TΨC arm stem, whereas three tRNAs (trnA, trnS1, and trnS2) lost the dihydrouracil (DHU) arm. The rrnL was located between trnL1 and trnV and the rrnS was determined between trnV and trnQ. The length of rrnL and rrnS is 1,020 bp and 697 bp, and their A + T content were 82.55% and 82.07%, respectively. The control region was 1,638 bp in length with an A + T content of 77.84% and located between the trnQ and trnM.
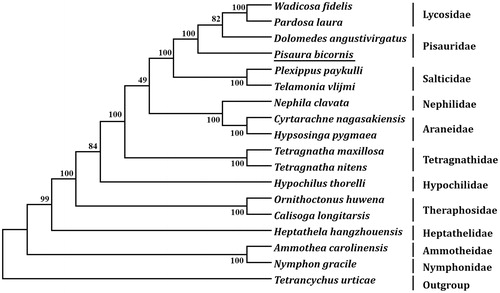
Eleven PCGs started with a typical ATN start codons (ATT and ATA), and the remaining two PCGs (cox1 and cox3) began with TTG. Ten PCGs terminated with TAA, two (nad1 and nad2) terminated with TAG, whereas nad4L terminated with an incomplete stop codon T. Based on the concatenated amino acid sequences of 13 PCGs, the neighbor-joining method was used to construct the phylogenetic relationship of P. bicornis with 16 other known spiders. The result showed that P. bicornis is closely related to Dolomedes angustivirgatus and belongs to Pisauridae ().
Figure 1. Phylogenetic tree showing the relationship between Pisaura bicornis and 16 other spiders based on neighbor-joining method. GenBank accession numbers used in the study are the following: Ammothea carolinensis (GU065293), Calisoga longitarsis (NC_010780), Cyrtarachne nagasakiensis (KR259802), Dolomedes angustivirgatus (NC_031355), Heptathela hangzhouensis (NC_005924), Hypochilus thorelli (EU523753), Hypsosinga pygmaea (KR259803), Nephila clavata (AY452691), Nymphon gracile (NC_008572), Ornithoctonus huwena (AY309259), Pardosa laura (KM272948), Pisaura bicornis (MN296112), Plexippus paykulli (KM114572), Telamonia vlijmi (NC_024287), Tetragnatha maxillosa (KP306789), Tetragnatha nitens (KP306790), Tetrancychus urticae (EU345430), and Wadicosa fidelis (NC_026123). T. urticae was used as an outgroup. Spider determined in this study was underlined.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication. The authors also are responsible for the content and writing of the paper.
Additional information
Funding
References
- Li C, Wang ZL, Fang WY, Yu XP. 2016. The complete mitochondrial genome of the orb-weaving spider Neoscona theisi (Walckenaer) (Araneae: Araneidae). Mitochondr DNA A DNA Mapp Seq Anal. 27:4035–4036.
- Platnick NI. 2015. The world spider catalog, Version 15.0. American Museum of Natural History. http://research.amnh.org/iz/spiders/catalog/.
- Sierwald P. 1997. Phylogenetic analysis of Pisaurine nursery web spiders, with revisions of Tetragonophthalma and Perenethis (Araneae, Lycosoidea, Pisauridae. J Arachnol. 25:361–407.
- Yang WJ, Xu KK, Liu Y, Yang DX, Li C. 2019. Complete mitochondrial genome and phylogenetic analysis of Argiope perforata (Araneae: Araneidae). Mitochondr DNA B. 4:1963–1964.
- Zhang JX. 2000. Taxonomy studies on Chinese spiders the genus Pisaura (Araneae: Pisauridae. Acta Arachnol Sin. 9:1–9.
