Abstract
We determined the complete mitogenome sequence of Melanotus cribricollis (Faldermann) (GenBank accession no. MK792748). The mitogenome is 15,908 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and one non-coding control region. The overall nucleotide composition was 40.8% A, 31.5% T, 17.4% C, and 10.3% G, with 72.3% of AT, respectively. Phylogenetic analysis based on the 13 coding protein genes nucleotide sequences revealed that M. cribricollis clustered with the same genus species M. villosus, and the three genus Melanotus Eschscholtz, Agriotes Eschscholtz and Adrastus Eschscholtz had a close relationship.
Elaterids are among the most abundant beetles in many terrestrial habitats and some in larval form are destructive agricultural pests. Elateridae commonly known as click beetles contains approximately 10,000 species (Johnson Citation2002). Melanotus cribricollis, black integrally, widely distributed in China and East Asia (Jiang and Wang Citation1999). The sampled specimen was collected from the city of Shangluo, China (the geospatial coordinates: 33°51′57″N, 109°57′05″E) in April 2018. The specimen was stored in the Entomological specimen room of Shangluo University (voucher no. CO-2018230). Genomic DNA was extracted from muscle tissues using DNeasy DNA Extraction kit (Qiagen, Hilden, Germany). The complete mitochondrial DNA sequence of M. cribricollis was determined by Illumina HiSeq 2500 Sequencing System. In total, 5.8 G raw reads were obtained, quality-trimmed, and assembled using MITObim v 1.7 (Hahn et al. Citation2013). The genome was annotated using software GENEIOUS R8 (Biomatters Ltd., Auckland, New Zealand).
The complete mitochondrial genome of M. cribricollis was 15,908 bp in total length and had been deposited in GenBank database with an accession number MK792748. The overall base composition was 40.8% A, 31.5% T, 17.4% C, and 10.3% G, with an A + T ratio of 72.3%. The gene arrangement of M. cribricollis is found to be similar to most insect mitochondrial genomes (Wolstenholme Citation1992). The full mitochondrial genome contains 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a putative control region (CR). Most PCGs of M. cribricollis have the conventional start codon for invertebrate mitochondrial PCGs (ATN), with the exception of nad4 (GTG), nad1 (TTG) and cox1 (AAT), as the asparagine (AAT or AAC) are proposed to be the start codon for cox1 in suborder Polyphaga (Sheffield et al. Citation2008). Most of the PCGs terminate with the stop codon TAA or TAG, whereas nad5, cox2, and cox3 end with the incomplete codon T. The 22 tRNA genes range from 63 bp (trnC) to 70 bp (trnK, trnW and trnV). Two rRNA genes (rrnL and rrnS) locate at trnL1/trnV and trnV/control regions, respectively. The lengths of the two rRNA genes (rrnL and rrnS) in M. cribricollis are about 1280 and 739 bp, with the A + T contents of 78.5 and 75.8%, respectively. The length of control region is 1281 bp with the AT content of this region is up to 79.8%.
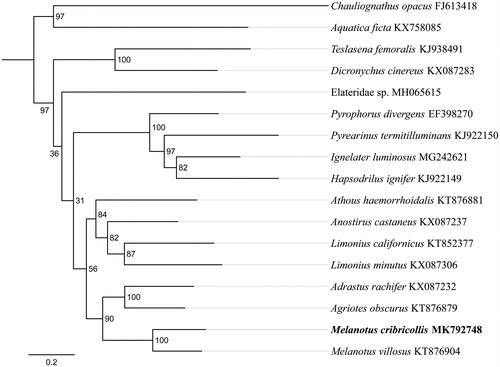
To validate the phylogenetic position of M. cribricollis, 13 mitochondrial protein-coding genes sequences were extracted from the complete mitochondrial DNA sequences of 15 closely related taxa of Elateridae. The phylogenetic tree was constructed using the maximum-likelihood method through raxmlGUI 1.5 (Silvestro and Michalak Citation2012). Results showed that the family Elateridae is monophyletic, which was consistent with the previous studies (Douglas Citation2011; Lin et al. Citation2018) (). The new sequenced species M. cribricollis got together with the same genus species Melanotus villosus, and Melanotus was sister to Agriotes and Adrastus, indicating the close relationship of these three genus. As conclusion, we obtained and described the complete mitochondrial genome of M. cribricollis, and the phylogenetic tree provided a reference for understanding the taxonomic status.
Disclosure statement
All authors have read and approved the final manuscript. No conflict of interest was reported by the authors.
Additional information
Funding
References
- Douglas H. 2011. Phylogenetic relationships of Elateridae inferred from adult morphology, with special reference to the position of Cardiophorinae. Zootaxa. 2900:1–45.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Jiang SH, Wang SY. 1999. Economic click beetle faun-na of China (Coleoptera: Elateridae). Beijing: China Agriculture Press (Chinese).
- Johnson PJ. 2002. Elateridae. Vol. 2. In: Arnett RH, Thomas MC, Skelley PE, Frank JH, editors. American Beetles. Boca Raton, Florida, USA: CRC Press LLC; p. 160–173.
- Lin A, Zhao X, Song N, Zhao T. 2018. Analysis of the complete mitochondrial genome of click beetle Agriotes hirayamai (Coleoptera: Elateridae). Mitochondr DNA B. 3:290–291.
- Sheffield N, Song H, Cameron S, Whiting M. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 25:2499–2509.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.