Abstract
Sargassum fusiforme is an important economic seaweed in East Asia. In this study, we characterized the complete chloroplast genome sequence of S. fusiforme using PacBio long-read sequencing technology. It had a circular mapping molecular with the length of 124,286 bp, with a large single-copy region (LSC, 73,437 bp) and a small single copy region (SSC, 40,131 bp) separated by a pair of inverted repeats (IRs, 5,359 bp). The cp genome contained 173 genes including 139 protein-coding, 6 rRNA, and 28 tRNA genes. The phylogenomic analysis indicated that S. fusiforme is closely related to S. thunbergii.
Sargassum fusiforme (Harvey) Setchell (= Hizikia fusiformis (Harvey) Okamura), widely applied as food, polysaccharide source, and medical agent, is a perennial brown macroalga distributed in the lower intertidal zones along the west of northern Pacific (Yu et al. Citation2012). Despite the importance of the species, there has been no genomic studies on S. fusiforme except the study of its mitochondrial genome (Liu et al. Citation2016). In this study, we reported and characterized the complete chloroplast genome of S. fusiforme using PacBio long-read sequencing technology. Moreover, the phylogeny of Phaeophyceae was reconstructed by utilizing the published related species’ chloroplast genome sequences to reassess the reliable taxonomic status of S. fusiforme.
One S. fusiforme individual was collected from Dongtou Island, Zhejiang Province of China (27°48′50′′ N, 121°10′1′′ E), and stored at Wenzhou university at −80 °C for DNAs isolation. Its voucher specimen was deposited in the Herbarium of Wenzhou University (Qian, qwg141101). Dozens of leaves (vesicle) were used to extract the total genomic DNA of S. fusiforme by using a plant genomic DNA extraction kit (Annoroad Gene Technology, Beijing, China). The chloroplast genome was reconstructed based on the PacBio Sequel data. Approximately 13-Gb sequence data were randomly extracted from the total sequencing output and used as input for Organelle_PBA (Soorni et al. Citation2017) to assemble the chloroplast genome. The chloroplast genome of S. thunbergii (GenBank accession number: NC_029134) was used as the reference sequence. The Illumina HiSeq data were used to correct the assembled genome using the Pilon software program (Walker et al. Citation2014). Gene annotation was conducted via the online program Dual Organellar Genome Annotator (DOGMA; Wyman et al. Citation2004) using the same method as Liu, Li, Worth, et al. (Citation2017) and Liu, Li, et al. (Citation2018).
The complete cp genome of S. fusiforme (GenBank accession number: MN121852) was 124,286 bp long comprising a pair of inverted repeat regions (IRs with 5,359 bp) divided by two single-copy regions (LSC with 73,437 bp and SSC with 40,131 bp). The overall GC content of the total length, LSC, SSC, and IR regions were 30.4%, 59.1%, 32.3%, and 8.6%, respectively. The cp genome encoded a total of 173 genes, of which 168 were unique and 5 were duplicated in the IR regions. The 173 unique genes consisted of 139 protein-coding genes, 28 tRNA genes, and 6 rRNA genes.
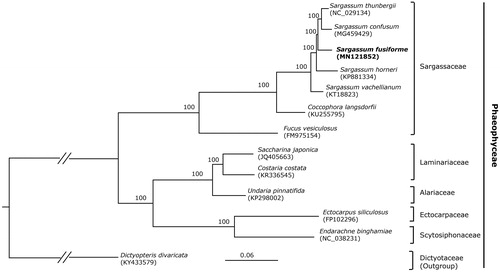
Maximum-likelihood (ML) analyses were run on a data set that included 126 protein-coding genes for 13 taxa in Phaeophyceae using RAxML v. 8.2.12 on CIPRES (http://www.phylo.org) under the GTR model. Our phylogenomic tree showed a good resolution of the species of Sargassaceae and other families in Phaeophyceae, with full support for all the nodes. The phylogenomic result () is consistent with the prior phylogenetic study on the taxonomic status of S. fusiforme (Stiger et al. Citation2003; Liu, Pang, et al. Citation2016; Liu, Pan, et al. Citation2018) confirming that S. fusiforme should not be assigned to the distinct genus Hizikia. The phylogenomic analysis showed that S. fusiforme is closely related to S. thunbergii.
Disclosure statement
The authors are really grateful to the opened raw genome data from public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Liu LX, Li P, Zhang HW, Worth J. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T. sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Forest Res. 23:318–323.
- Liu F, Pan J, Zhang ZS, Moejes FW. 2018. Organelle genomes of Sargassum confusum (Fucales, Phaeophyceae): mtDNA vs cpDNA. J Appl Phycol. 30:2715–2722.
- Liu F, Pang SJ, Luo MB. 2016. Complete mitochondrial genome of the brown alga Sargassum fusiforme (Sargassaceae, Phaeophyceae): genome architecture and taxonomic consideration. Mitochondrial DNA. 27:1158–1160.
- Soorni A, Haak D, Zaitlin D, Bombarely A. 2017. Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data. BMC Genomics. 18:49.
- Stiger V, Horiguchi T, Yoshida T, Coleman AW, Masuda M. 2003. Phylogenetic relationships within the genus Sargassum (Fucales, Phaeophyceae), inferred from ITS-2 nrDNA, with an emphasis on the taxonomic subdivision of the genus. Phycological Res. 51:1–10.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLos One. 9:e112963.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yu SH, Deng YY, Yao JT, Li SY, Xin X, Duan DL. 2012. Population genetics of wild Hizikia fusiformis (Sargassaceae, Phaeophyta) along China’s coast. J Appl Phycol. 24:1287–1294.