Abstract
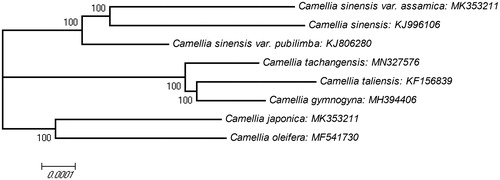
To understand genetic background and phylogenetic position of Camellia tachangensis, we determined its complete chloroplast genome sequence which is 157,026 bp in length with overall GC content of 36.7%. It has four sub regions: a large single-copy (LSC) region (86,669 bp) and a small single-copy (SSC) region (18,253 bp) are separated by two inverted repeat (IR) regions (26,052 bp each). A total of 129 genes were annotated, containing 86 protein-coding genes, 35 tRNA genes, and 8 rRNA genes. Phylogenetic trees showed C. tachangensis clustered with Camellia gymnogyna and Camellia taliensis and separated from Camellia sinensis and its two varieties, Camellia sinensis var. assamica and Camellia sinensis var. pubilimba.
Camellia is the economically most important genus in the family Theaceae. It contains more than 200 species and is regarded as one of the most difficult taxa in plants (Vijayan et al. Citation2009). Until now, taxonomists still have different opinions on the taxa of section Thea of the genus Camellia. The chloroplast genome sequences could provide valuable information for taxonomic classification and the reconstruction of phylogeny for resolving complex evolutionary relationships (Jansen et al. Citation2007; Parks et al. Citation2009). In the past few years, the complete chloroplast genome sequences of most species in section Thea were determined (Yang et al. Citation2013; Chen et al. Citation2014; Huang et al. Citation2014; Dong et al. Citation2018; Li et al. Citation2018; Zeng et al. Citation2018). However, the complete chloroplast genome sequence of Camellia tachangensis F. C. Zhang has not been reported. In this study, we characterized the complete chloroplast genome sequence of C. tachangensis as a resource for better understanding the diversification and evolution of the genus Camellia.
The young leaves were collected from C. tachangensis cultivar Xingyi6, growing in the China National Germplasm Hangzhou Tea Repository (30.179584N, 120.093516E). A voucher specimen (GS00125) was deposited in the Herbarium of the Germplasm Resources Laboratory of the Tea Research Institute of the Chinese Academy of Agricultural Sciences. The DNA sequencing was performed on an Illumina Hiseq 2500 platform (Illumina, San Diego, CA). The chloroplast genome was de novo assembled using NOVOPlasty (Dierckxsens et al. Citation2017) with the rbcL gene sequence (Genbank: MH270471.1) as seed and annotated using GeSeq (Tillich et al. Citation2017).
The complete chloroplast genome of ‘Xingyi6’ (Genbank: MN327576) is 157,026 bp in length with the overall GC ratio of 36.7%. It composed of a large single-copy (LSC) region of 86,669 bp and a small single-copy (SSC) region of 18,253 bp separated by two inverted repeat (IR) regions of 26,052 bp each. A total of 129 genes were annotated, containing 86 protein-coding genes, 35 tRNA genes, and 8 rRNA genes. Among them, four rRNA genes (i.e. 4.5S, 5S, 16S, and 23S rRNA), seven protein-coding genes (i.e. ndhB, rpl2, rpl23, rps12, rps7, ycf15, ycf1, and ycf2), and seven tRNA genes (i.e. trnM, trnL, trnV, trnE, trnA, trnR, and trnN) occur in double copies in IR regions. There are two protein-coding genes (clpP and rps12) containing two introns and 11 protein-coding genes (atpF, ndhA, ndhB × 2, petB, petD, rpl2 × 2, ropC1, rps16, and ycf3) containing one intron.
Using clustalw2.1, we aligned eight chloroplast genomes of different species from the genus Camellia. A molecular phylogenetic tree was constructed with the MEGA X (Kumar et al. Citation2018) using neighbour-joining (NJ) method with 10,000 bootstrap replicates (). The result showed C. tachangensis, Camellia gymnogyna, and Camellia taliensis were clustered into a group. Camellia sinensis and its two varieties, Camellia sinensis var. assamica and Camellia sinensis var. pubilimba were clustered into another group. This chloroplast genome will provide valuable information for genetic evolution studies of Camellia.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen CM, Ma CL, Ma JQ, Liu SC, Chen L. 2014. Sequencing of chloroplast genome of Camellia sinensis and genetic relationship for Camellia plants based on chloroplast DNA sequences. J Tea Sci. 34:371–380.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Dong M, Liu SQ, Xu ZG, Hu ZY, Ku WZ, Wu L. 2018. The complete chloroplast genome of an economic plant, Camellia sinensis cultivar Anhua, China. Mitochondrial DNA Part B: Resour. 3:558–559.
- Huang H, Shi C, Liu Y, Mao SY, Gao LZ. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14:151.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci. 104:19369–19374.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: *Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li WX, Xing F, Ng WL, Zhou YB, Shi XG. 2018. The complete chloroplast genome sequence of Camellia ptilophylla (Theaceae): a natural caffeine-free tea plant endemic to China. Mitochondrial DNA Part B Resour. 3:426–427.
- Parks M, Cronn R, Liston A. 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7:84.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Vijayan K, Zhang WJ, Tsou CH. 2009. Molecular taxonomy of Camellia (Theaceae) inferred from nrITS sequences. Am J Bot. 96:1348–1360.
- Yang JB, Yang SX, Li HT, Yang J, Li DZ. 2013. Comparative chloroplast genomes of Camellia species. PLoS One. 8:e73053.
- Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, Yang JB. 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods. 14:43.