Abstract
Larvae of click beetles are the most important soil-dwelling agricultural pests and are abundant throughout the world. Agrypnus sp. belongs to the subfamily Agrypninae of Elateridae. Here, we sequenced and annotated the complete mitochondrial genome (mitogenome) of Agrypnus sp., the new representative of the mitogenome of the subfamily. This mitogenome was 16,056 bp long and encoded 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA unit genes (rRNAs). Gene order was conserved and identical to most other previously sequenced Elateridae. The nucleotide composition biases towards A and T, which together made up 70.8% of the entirety. Phylogenetic analysis based on 13 PCGs sequences showed that Agrypnus sp. got together with the same subfamily species Pyrophorus divergens, Pyrearinus termitilluminans, Ignelater luminosus, and Hapsodrilus ignifer with high support value.
Click beetles (Coleoptera: Elateridae) are the largest family of Elateroidea and include approximately 9000 species. Species in this group often exhibit a unique and well-known startling defense mechanism called ‘clicking’ (Sagegami-Oba et al. Citation2007). Wireworms, the larvae of click beetles, are the most important soil-dwelling agricultural pests worldwide and can damage potato, carrot, cereals, sugar beet, sugarcane, and soft fruits (Toth et al. Citation2003; Barsics et al. Citation2013).
The specimens of Agrypnus sp. used for this study were collected from Ledong County, Hainan Province, China (18°46′N, 108°53′E, May 2017) and were stored in Entomological Museum of Gannan Normal University (Accession number GNU-AGRY041). After morphological identification, total genomic DNA was extracted from tissues using DNeasy DNA Extraction kit (Qiagen, Hilden, Germany ). Mitogenome sequence was generated using Illumina HiSeq 2500 Sequencing System. In total, 6.5 G raw reads were obtained, quality-trimmed, and assembled using MITObim v 1.7 (Hahn et al. Citation2013). By comparison with the homologous sequences of other Elateridae species from GenBank, the mitogenome of Agrypnus sp. was annotated using software GENEIOUS R8 (Biomatters Ltd., Auckland, New Zealand).
The complete mitogenome of Agrypnus sp. is 16,056 bp (Genbank accession, MN370897). It contains 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and 1 non-coding AT-rich region. The nucleotide composition of the mitogenome was biased toward A and T, with 70.8% of A + T content (A 40.2%, T 30.6%, C 18.6%, G 10.6%). Gene order was conserved and identical to that of Drosophila yakuba and to most other previously sequenced Elateridae (Amaral et al. Citation2016; Linard et al. Citation2016; Wang et al. Citation2019). Four PCGs (nad4, nad4l, nad5, and nad1) were encoded by the minority strand (N-strand) while the other nine were located on the majority strand (J-strand). Most PCGs of Agrypnus sp. have the conventional start codons ATN (five ATG, three ATA, and three ATT), with the exception of nad1 (TTG) and cox1 (AAC), as the asparagine (AAT or AAC) are proposed to be the start codon for cox1 in suborder Polyphaga (Sheffield et al. Citation2008). Except for four genes (cox2, cox3, nad5, and nad4) end with the incomplete stop codon T−, all other PCGs terminated with the stop codon TAA or TAG. The 22 tRNA genes vary from 62 bp (trnC) to 70 bp (trnK and trnV). Two rRNA genes (rrnL and rrnS) locate at trnL1/trnV and trnV/control region, respectively. The lengths of rrnL and rrnS in Agrypnus sp. are 1281 and 736 bp, with the AT contents of 75.3 and 73.5%, respectively.
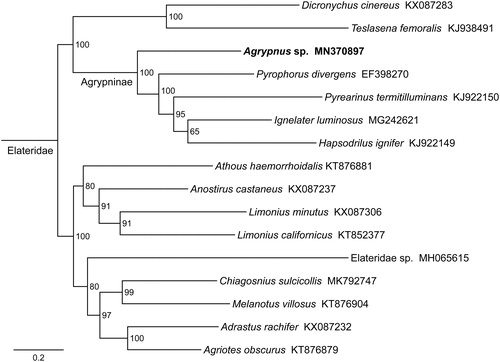
Phylogenetic tree was constructed using the maximum-likelihood method through raxmlGUI 1.5 (Silvestro and Michalak Citation2012) based on 13 mitochondrial protein-coding genes sequences. Results showed that the newly sequenced species Agrypnus sp. got together with the same subfamily Agrypninae species Pyrophorus divergens, Pyrearinus termitilluminans, Ignelater luminosus, and Hapsodrilus ignifer with high support value (). In conclusion, the mitogenome of Agrypnus sp. is obtained in this study and can provide essential and important DNA molecular data for further phylogenetic and evolutionary analysis of Elateridae.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Amaral DT, Mitani Y, Ohmiya Y, Viviani VR. 2016. Organization and comparative analysis of the mitochondrial genomes of bioluminescent Elateroidea (Coleoptera: Polyphaga). Gene. 586:254–262.
- Barsics F, Haubruge E, Verheggen F. 2013. Wireworms’ management: an overview of the existing methods, with particular regards to Agriotes spp. (Coleoptera: Elateridae). Insects. 4:117–152.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Linard B, Arribas P, Andújar C, Crampton‐Platt A, Vogler AP. 2016. Lessons from genome skimming of arthropod-preserving ethanol. Mol Ecol Resour. 16:1365–1377.
- Sagegami-Oba R, Oba Y, Ôhira H. 2007. Phylogenetic relationships of click beetles (Coleoptera: Elateridae) inferred from 28S ribosomal DNA: insights into the evolution of bioluminescence in Elateridae. Mol Phylogenet Evol. 42:410–421.
- Sheffield N, Song H, Cameron S, Whiting M. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 25:2499–2509.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Toth M, Furlan L, Yatsynin VG, Ujvary I, Szarukan I, Imrei Z, Tolasch T, Francke W, Jossi W. 2003. Identification of pheromones and optimization of bait composition for click beetle pests (Coleoptera: Elateridae) in Central and Western Europe. Pest Manag Sci. 59:417–425.
- Wang Y, Wang X, Liu Y. 2019. The complete mitochondrial genome of click beetle Chiagosnius sulcicollis (Coleoptera: Elateridae) and phylogenetic analysis. Mitochondr DNA B. 4:2324–2325.