Abstract
The Arctic moss Aulacomnium turgidum (Wahlenb.) Schwaegr. is distributed widely above the Arctic Circle and can regenerate successfully after 400 years of ice entombment. Here, we report the complete mitogenome sequence of A. turgidum (103,937 bp). The genome contains 3 ribosomal RNAs, 24 transfer RNAs, and 40 protein-encoding genes. In a phylogenetic tree generated using the combined amino acid sequences of 32 mitochondrial genes from A. turgidum, 25 Bryophyta, and three Marchantiophyta, the phylogenetic position of A. turgidum (Rhizogoniales) is close to that of the Hypnales and Ptychomniales, forming a monophyletic clade with perfect supporting values.
Svalbard, a Norwegian archipelago, has a typical Arctic environment. Meteorological records for the period 1993–2011 indicate that the annual mean surface air temperatures in this region vary between 2.5 and 5.8 °C in summer and −11.4 and −9.4 °C in winter (Maturilli et al. Citation2013). Despite the harsh temperatures, plants with a high tolerance of abiotic stresses, including bryophytes, are widely distributed in this region (Crawford et al. Citation1994; Liu et al. Citation2010). Of these, the moss Aulacomnium turgidum (Wahlenb.) Schwaegr. is widely distributed above the Arctic Circle, including Svalbard, Greenland, and Alaska (GBIF Secretariat Citation2017). Because A. turgidum can regenerate following 400 years of ice entombment, we postulated that it has very high freezing stress tolerance (La Farge et al. Citation2013). In this study, we analyzed the mitogenome of A. turgidum and conducted a molecular phylogenetic analysis of bryophytes.
Specimens of A. turgidum growing under natural conditions were collected near the Korean Dasan Arctic Station (78°55′27′′ N; 11°56′34′′ E) in Ny-Ålesund, Svalbard in August 2006. They were placed in plastic containers and transported to Korea, where they were cultivated on BCD solid medium (Ashton and Cove Citation1977) in a growth room at 25 °C with continuous light. A dried specimen was deposited in the Korea Polar Research Institute (KOPRI) Herbarium (https://kvh.kopri.re.kr) under the number KOPRI-MO00899.
Total genomic DNA was extracted from 100 μg of a fresh sample of A. turgidum and used to prepare the library through TruSeq DNA Sample Prep Kit (Illumina, San Diego, CA). Paired-ends reads were sequenced using Illumina HiSeq2500 at 2 × 101 bp (Illumina, San Diego, CA) and de novo assembled using CLC Genomics Workbench V7.5 (CLC Bio, Aarhus, Denmark) as described previously (Cho et al. Citation2019). All contigs were blasted to the Physcomitrella patens mitogenome (NC_007945), and contigs with highest identities were selected, joined via serial raw reads mapping processes. The assembled mitogenome sequences were annotated using GeSeq (Tillich et al. Citation2017) and manually curated with BLAST searches.
Mitogenome of A. turgidum (GenBank accession number: MN233806) has a total length of 103,937 bp (48× average coverage). Its GC ratio is 41.1%, similar to that of P. patens (40.6%) and it contains 3 ribosomal RNA, 24 transfer RNA, and 40 protein-coding genes. The mitochondrial structure and gene order are similar to those of other bryophytes.
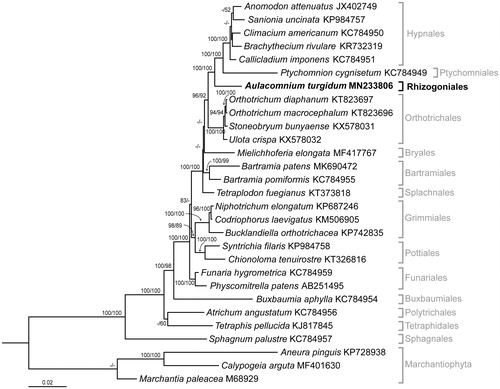
Phylogenetic analyses of A. turgidum were performed using the amino acid sequences of 32 genes (8094 amino acids in length) from 28 mitogenomes of 25 Bryophyta and three Marchantiophyta, and were aligned using MAFFT ver. 7 (Katoh and Standley Citation2013). Maximum-likelihood (ML) and neighbor-joining (NJ) trees with 1000 bootstrap replicates were constructed in MEGA7 (Kumar et al. Citation2016). The phylogenetic position of A. turgidum (Rhizogoniales) was found to be close to those of Hypnales and Ptychomniales, and these species formed a monophyletic clade with perfect bootstrap support (). To date, over 60 mitogenomes from Bryophyta have been reported, but this is the first report from the order Rhizogoniales. The A. turgidum mitogenome sequence will be useful for evolutionary studies of bryophytes, including Arctic species.
Figure 1. Maximum-likelihood (ML) tree inferred from the amino acid sequences of 32 mitochondrial genes with JTT + G + F parameters in MEGA7. ML and NJ trees were constructed using 1000 bootstrap replicates. Bootstrap values above 80 and 50 are shown on the branches of the ML and NJ trees, respectively.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Ashton NW, Cove DJ. 1977. The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Molec Gen Genet. 154:87–95.
- Cho SM, Byun MY, Lee H, Park H, Lee J. 2019. The complete mitogenome of the Antarctic moss Bartramia patens Brid. Mitochondrial DNA Part B. 4:1759–1760.
- Crawford RM, Chapman HM, Hodge H. 1994. Anoxia tolerance in high Arctic vegetation. Arct Alp Res. 26:308–312.
- GBIF Secretariat. 2019. GBIF Backbone Taxonomy. Checklist dataset; [accessed 2019 Aug 20]. 10.15468/39omei
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- La Farge C, Williams KH, England JH. 2013. Regeneration of little ice age bryophytes emerging from a polar glacier with implications of totipotency in extreme environments. Proc Natl Acad Sci USA. 110:9839–9844.
- Liu S, Lee H, Kang P-S, Huang X, Yim JH, Lee HK, Kim I-C. 2010. Complementary DNA library construction and expressed sequence tag analysis of an Arctic moss, Aulacomnium turgidum. Polar Biol. 33:617–626.
- Maturilli M, Herber A, König-Langlo G. 2013. Climatology and time series of surface meteorology in Ny-Ålesund, Svalbard. Earth Syst Sci Data. 5:155–163.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
