Abstract
One new mitochondrial genome of Indonemoura auriformis from the family Nemouridae (Insecta: Plecoptera) was sequenced in the study. The mitochondrial genome has the length of 15,718 bp, encoding 37 genes: 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 ribosomal RNA (rRNA) genes. The whole nucleotide composition biased adenine and thymine with A + T accounting for 69.9%. Nine PCGs and 14 tRNA genes are encoded in the J chain, the other four PCGs, eight tRNAs, and two rRNA genes are encoded in the chain of N. The mitochondrial genome includes 13 gene overlaps and 12 intergenic spacers. Most PCGs strictly use the ATN as start codon, and terminate with traditional stop codon (TAA and TAG). Except the tRNASer(AGN) seems to lack dihydrogen glycine arm, all tRNA genes of the mitochondrial genome are the typical clover secondary structure. The phylogenetic tree of PCGs dataset based on bayesian inference (BI) and maximum likelihood (ML) analysis show the same tree topology. Both ML and BI analysis support the sister-group relationship between Amphinemuria and Nemoura. Meanwhile, Capniidae is closely related to Taeniopterygidae.
The mitochondrial genome of insect usually contains 13 protein-coding (PCG), 22 transfer RNA (tRNA), 2 ribosomal RNA (rRNA) gene, and an assumed control region (Lin and Danforth Citation2004; Gissi et al. Citation2008). The Nemouridae is of important systematic status and ecological importance, more than 1100 species of this family have been discovered (DeWalt et al. Citation2019), which are distributed in aquatic areas all over the world except the Arctic continent. To date, two mitochondrial genomes from the Genus Indonemoura have been reported in GenBank or registered (Cao et al. Citation2019). To facilitate a more comprehensive study of mitogenome phylogeny in the Plecoptera, we analyzed the mitogenome of Indonemoura auriformis and reconstructed the phylogeny of Nemouroidea. Adult specimens of I. auriformis were collected from Nanping, Henan Province, China, by Weihai Li and Jinjun Cao in June 2017 (35.712°N, 113.620°E). We preserved specimens in 100% ethanol in the field and stored them at –20 °C until tissues were used for DNA extraction. A voucher specimen of I. auriformis (No. VHL-0009) was stored in the Entomological Museum of the Henan Institute of Science and Technology (HIST), Henan Province, China. We extracted total genomic DNA from thoracic muscle tissue by using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
The entire circular genome has the length of 15,718 bp (GenBank no: MN419915) and contains 37 genes and a control region. Like previous published stoneflies, 23 genes encode most coding chains (J chains) and 14 genes encode a few coding chains (N chains). The entire genome accounts for 69.90% of the base composition, including 13 gene overlaps and 12 intergenic spacers. Most PCG genes initiated using the typical starting codon ATN, but TTG is recommended as the starting codon for ND1. Eleven PCG genes use TAA or TAG as stop codon, two genes (COII and ND5) terminated with single T. The length of each tRNA ranged from 63 to 71 bp, most tRNAs can be folded into a typical clover structure except the dihydrodoladine arm of tRNASer(AGN) forms a simple loop. The A + T-rich region is located between srRNA and tRNAIle with the length of 852 bp. This region is considered to be the control region because it contains sources of transcription and replication (Taanman Citation1999; Cameron Citation2014).
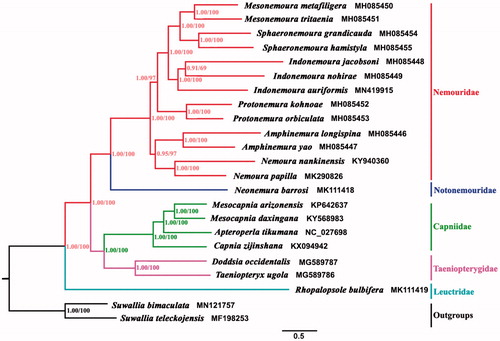
Phylogenetic trees from 21 Nemouroidea species which belong to 5 families were reconstructed using published DNA data, and two species (Suwallia bimaculata and Suwallia teleckojensis) from Chloroperlidae were outgroups, as shown in . The phylogenetic tree conducted from PCGs dataset based on Bayesian inference (BI) and Maximum likelihood (ML) analysis provide the same tree topology. In Nemouridae, the ML and BI trees both support the sister-group relationship between Amphinemura and Nemoura, and the monophyletic clade of genus Indonemoura. Capniidae is closely related to Taeniopterygidae. Meanwhile, the position of Leuctridae is consistent with the previous traditional morphological classification (Illies Citation1965; Zwick Citation2000, Citation2009). This study needs the mitochondrial genome data to understand the phylogeny of Nemouroidea.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Cao JJ, Wang Y, Li WH. 2019. Comparative mitogenomic analysis of species in the subfamily Amphinemurinae (Plecoptera: Nemouridae) reveal conserved mitochondrial genome organization. Int J Biol Macromol. 138:292–301.
- DeWalt RE, Maehr MD, Neu-Becker U, Stueber G. 2019. Plecoptera species file online. http://Plecoptera.SpeciesFile.org; [accessed 2019 Sep 15].
- Gissi C, Iannelli F, Pesole G. 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 101:301–320.
- Illies J. 1965. Phylogeny and zoogeography of the Plecoptera. Annu Rev Entomol. 10:117–140.
- Lin CP, Danforth BN. 2004. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phylogenet Evol. 30:686–702.
- Taanman JW. 1999. The mitochondrial genome: structure, transcription, translation and replication. BBA: Biomembranes. 1410:103–123.
- Zwick P. 2000. Phylogenetic system and zoogeography of the Plecoptera. Annu Rev Entomol. 45:709–746.
- Zwick P. 2009. The Plecoptera – who are they? The problematic placement of stoneflies in the phylogenetic system of insects. Aquatic Insects. 31:181–194.