Abstract
Eriobotrya fragrans Champion ex Bentham is a potential medicinal plant of the genus Eriobotrya Lindl in the family Rosaceae. To better determine its phylogenetic location with respect to the other Rosaceae species, the complete chloroplast genome of E. fragrans was sequenced. The whole E. fragrans chloroplast genome is 159,286 bp in length, consisting of a pair of inverted repeat (IR) regions of 26,343 bp, one large single-copy (LSC) region of 87,301 bp, and one small single-copy (SSC) region of 19,299 bp. The overall GC content of the whole chloroplast genome is 36.7%. Further, phylogenetic analysis using maximum likelihood with TVM + F+R2 model strongly supports the relationship: sisterhood of E. fragrans and E. japonica, followed by three species of Pyrus L.
Eriobotrya fragrans Champ ex Bentham, an evergreen shrub tree species, is widely distributed in Guangdong, Guangxi, and Tibet of South China (http://foc.iplant.cn/). Among Eriobotrya Lindl plants, E. fragrans owns the stronger photosynthesis ability than the other species (Lin Citation2007). Eriobotrya fragrans has been reported to contain secondary metabolite polyphenols and flavonoids with antimicroorganism and antioxdant activities (Hong et al. Citation2008, Citation2009; Lin et al. Citation2011). For a better understanding of the relationships of E. fragrans and other Rosaceae species, it is necessary to perform high-throughput sequencing approaches.
About four gram fresh young and healthy leaves of E. fragrans in Mengla County (Yunnan, China; Long. 101.2546 E, Lat. 21.9263N, 564 m) were picked for DNA extraction from modified CTAB method (Liu et al. Citation2005). The voucher was deposited at the Key Laboratory of Forest Resources Conservation and Utilization in the Southwest Mountains of China Ministry of Education, Southwest Forestry University (Accession no. SWFU-SY35360). The whole chloroplast genome was sequenced following Yang et al. (Citation2014), and the long-range PCR was used for next-generation sequencing with nine pairs of universal primers. Taking the E. japonica as a reference (Huang Citation2019), the complete chloroplast genome of E. fragrans was assembled using GetOrganelle software (Jin et al. Citation2018), and the chloroplast genome was annotated in Geneious 8.1.3 (Biomatters Ltd, Auckland, New Zealand).
The E. fragrans chloroplast genome, with a length of 159,286 bp, was 149 bp larger than that of E. japonica (159,137 bp, KT633951). It was also 782 bp and 871 bp smaller than that of Malus domestica (160,068 bp, KY818915) and Pyrus ussuriensis (160,157 bp, MK172841). The chloroplast genome has the usual quadripartite structure, featuring a LSC region (large single-copy region 87,301 bp), a SSC region (small single-copy region 19,299 bp), and a pair of IR (inverted repeats 26,343 bp). The overall GC content is 36.7% (LSC, 34.5%; SSC, 30.3%; IR, 42.7%). The E. fragrans chloroplast genome encodes 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
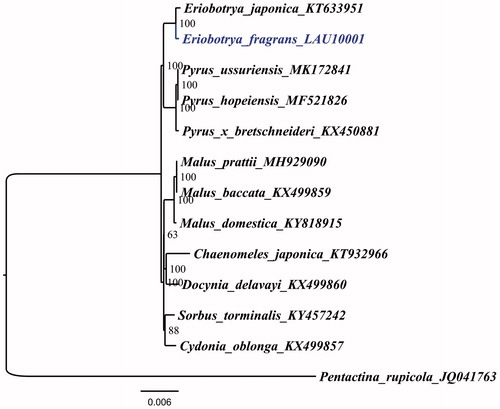
To determine the phylogenetic location of E. fragrans with respect to the other Rosaceae with fully sequenced chloroplast genomes, the complete E. fragrans chloroplast was used to reconstruct the phylogenetic relationships. With the chloroplast of Pentactina rupicola (Maleae, Rosaceae, JQ041763) as an out-group, 13 chloroplast genome sequences of Rosaceae, including Chaenomeles japonica (KT932966), Cydonia oblonga (KX499857), Docynia delavayi (KX499860), E. fragrans, E. japonica, Mulas baccatawere (KX499859), M. domestica, M. prattii (MH929090), Pyrus hopeiensis (MF521826), P. ussuriensis, P. x bretschneideri (KX450881), and Sorbus torminalis (KY457242), aligned by the MAFFT version 7 programme (Katoh and Standley Citation2013). A maximum-likelihood analysis based on the TVM + F+R2 model was performed with iqtree version 1.6.7 using 1000 bootstrap replicates (Nguyen et al. Citation2015). The phylogenetic tree reveals that E. fragrans and E. japonica is most closely related to Pyrus ussuriensis, P. hopeiensis, and P. x bretschneideri (), which is in agreement with previous reports on the close relationships between the two genera (Xiang et al. Citation2019).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data archiving statement
The chloroplast data of the E. fragrans will be submitted to Rosaceae Chloroplast Genome Database (https://lcgdb.wordpress.com). Accession numbers are LAU10001.
Additional information
Funding
References
- Hong YP, Huang SH, Cao HY, Lin SQ, Liu ZL. 2009. The comparison of antioxidant activity and the contents of antioxidant compounds of extract fractions between fragrant loquat and common loquat. Acta Hortic Sin. 36:898–904.
- Hong YP, Qiao YC, Lin SQ, Jiang YM, Chen F. 2008. Characterization of antioxidant compounds in Eriobotrya fragrans Champ leaf. Sci Hortic. 118(4):288–292.
- Huang J. 2019. Characterization of the complete chloroplast genome of Eriobotrya japonicain China and phylogenetic relationships. Mitochondrial DNA Part B. 4:1367–1369.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. DOI: 10.1101/256479.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lin BS, Cao HY, Hong YP. 2011. The comparison of antimicroorganism and antioxidant activity of leaf extract between fragrant loquat and common loquat. Food Sci Technol. 36:172–175.
- Lin SQ. 2007. World loquat production and research with special reference to China. Acta Hortic. 750:37–44.
- Liu YX, Yang XH, Lin SQ, Hu GB, Liu CM. 2005. An improved procedure for nuclear DNA isolation from Eriobotrya plants and its application. J Fruit Sci. 22:182–185.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Xiang YZ, Huang CH, Hu Y, Wen J, Li SS, Yi TS, Chen HY, Xiang J, Ma H. 2019. Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol Biol Evol. 34:262–281.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.