Abstract
The two complete mitochondrial genomes were sequenced from the Netherlands strain of the freshwater monogonont rotifer Brachionus calyciflorus. The mitochondrial genome sequences were 27,698 bp and 9,906 bp in size, respectively. The gene order and contents of the two B. calyciflorus strains were mostly identical to one another, except for the additional identification and translocation of several tRNAs in mitochondrial DNA I and II. Of 13 protein-coding genes (PCGs), three genes (ND1, ND5, and ND3) had incomplete stop codons. Furthermore, the start codon of ND2, CO2, and CO3 and ND4 genes was ATT, GTG, and ATA, respectively, while the start codon of other PCGs was ATG. The base composition of 13 PCGs of B. calyciflorus (the Netherlands strain) mitogenome showed 31.1% for A, 37.6% for T, 16.5% for C, and 14.8% for G, respectively.
In the freshwater rotifer Brachionus calyciflorus species complex, four and eight cryptic species were identified from Netherlands (Papakostas et al. Citation2016; Michaloudi et al. Citation2018) and China (Xiang et al. Citation2011), respectively, as Brachionus plicatilis species complex were identified from 15 species (Mills et al. Citation2017). However, to date, only one complete mitochondrial genome of B. calyciflorus (China) has been published (Nie et al. Citation2016), while several complete mitochondrial genome of other brackish Brachionus rotifers have been published; B. plicatilis (Suga et al. Citation2008), B. koreanus (Hwang et al. Citation2013; Hwang et al. Citation2014), and B. rotundiformis (Kim et al. Citation2017). Thus, the identification of B. calyciflorus species complex can be important to better understand the phylogenetic relationship of the freshwater rotifer B. calyciflorus species complex clade. Also, recently B. calyciflorus is considered as a model for environmental toxicology in response to environmental stressors (Zhang et al. Citation2015; Nys et al. Citation2016; Paraskevopoulou et al. Citation2018; Kim et al. Citation2018; Lee et al. Citation2019). The analysis of B. calyciflorus mitochondrial genome is important to identify and compare the species-specificity of the field-sampled and laboratory stocks. In this study, we identified two complete mitochondrial genomes of the monogonont rotifer B. calyciflorus (the Netherlands).
The resting eggs of B. calyciflorus were collected from sediments of freshwater ponds at in November 2015 (kindly provided by Dr. Steven A.J. Declerck, Netherlands Institute of Ecology, the Netherlands) in Zwartenhoek, the Netherlands (52°02′63″N and 4°18′35.5″E) and maintained at the Laboratory of Professor Atsushi Hagiwara, Nagasaki University in Japan. The type was deposited in the Ichthyological collection of the Faculty of Fisheries, Nagasaki University (FFNU) under the accession no. FFNU-Rot-0005. We sequenced 500 bp paired end library of B. calyciflorus from whole body genomic DNA using the Illumina HiSeq 2500 platform (GenomeAnalyzer, Illumina, San Diego, CA). De novo assembly was conducted by Newbler (version 2.9; identity 98) (http://www.454.com). Of the assembled B. calyciflorus 140,587 contigs, 14 mitochondrial contigs were obtained. After a manual curation of 14 contigs with Consed (version 19.0) (http://www.phrap.org/consed/consed.html), two contigs were finally obtained to the mitochondrial DNAs of B. calyciflorus.
The complete mitochondrial genomes of B. calyciflorus the Netherlands strain were 27,698 bp (mitochondrial DNA I; GenBank no. MN457951) and 9,906 bp (mitochondrial DNA II; GenBank no. MN457952) in size. The direction of 13 protein-coding genes (PGCs) of B. calyciflorus was identical to those of B. calyciflorus China strain (Nie et al., Citation2016). Of 13 protein-coding genes (PCGs), three genes (ND1, ND5, and ND3) had incomplete stop codons. Furthermore, the start codon of ND2, CO2, and CO3 and ND4 genes was ATT, GTG, and ATA, respectively, while the start codon of other PCGs was ATG. The base composition of 13 PCGs in the Netherlands strain of B. calyciflorus mitogenome showed 31.1% for A, 37.6% for T, 16.5% for C, and 14.8% for G, respectively. The mitochondrial genome A + T base composition (72.0%) of 13 PCGs was higher than G + C (28.0%), whereas the complete mitochondrial genome A + T base composition (68.7%) was higher than G + C (31.3%).
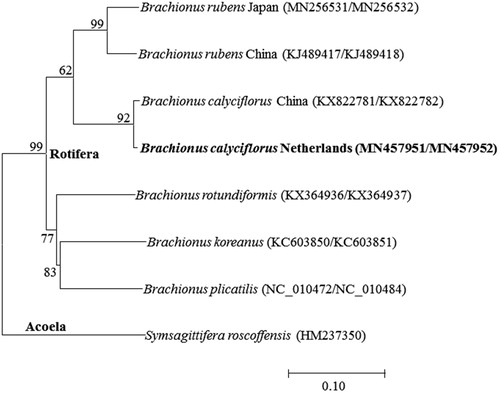
The placement of B. calyciflorus in the genus Brachionus with 13 PGCs was shown in . Two strains of B. calyciflorus were clustered closely to the freshwater rotifer Brachionus rubens. The gene order and contents of 13 PGCs of both B. calyciflorus were identical but tRNA-Ala and tRNA-Cys were rearranged in the mitochondrial DNA I, respectively. Particularly, tRNA-Leu and 16S rRNA were additionally located in the mitochondrial DNA I in B. calyciflorus (the Netherlands strain), compared to China strain, while B. calyciflorus China strain had nine extra tRNAs (e.g. tRNA-Ala and tRNA-Arg for mitochondrial DNA I and tRNA-Met, tRNA-Tyr, tRNA-Val, tRNA-Glu, tRNA-Trp, tRNA-His, and tRNA-Pro for mitochondrial DNA II), compared to the Netherlands strain. This indicates that the rearrangement and additional copies of tRNAs is likely occurring in sporadic manner in the genus B. calyciflorus species complex.
Figure 1. Phlyogenetic analysis of mitochondrial DNA. We conducted a comparison of the 13 mitochondiral DNA genes of Acoela and Rotifera. The 13 mitochondrial DNA genes were aligned by ClustalW. Maximum-likelihood analysis was performed by Mega software (ver. 10.0.1) with LG + G + I model. The rapid bootstrap analysis was conducted with 1000 replications with 48 threads running in parallel. The Acoela served as outgroup. -Ln = 24394.24.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Hwang D-S, Dahms H-U, Park HG, Lee J-S. 2013. A new intertidal Brachionus and intrageneric phylogenetic relationship among Brachionus as revealed by allometry and CO1-ITS1 gene analysis. Zool Stud. 52(1):13.
- Hwang D-S, Suga K, Sakakura Y, Hagiwara A, Park HG, Rhee J-S, Lee J-S. 2014. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera: Brachionidae). Mito DNA. 25(1):29–30.
- Kim H-S, Hwang D-S, Kim H-J, Sakakura Y, Hagiwara A, Lee J-S. 2017. Complete mitochondrial genome of the monogonont rotifer Brachionus rotundiformis (Rotifera, Brachionidae). Mito DNA B. 2(1):39–40.
- Kim H-S, Lee B-Y, Han J, Jeong C-B, Hwang D-S, Lee M-C, Kang H-M, Kim D-H, Kim H-J, Papakostas S, et al. 2018. The genome of the freshwater monogonont rotifer Brachionus calyciflorus. Mol Ecol Resour. 18(3):646–655.
- Lee J-S, Kang H-M, Jeong C-B, Han J, Park HG, Lee J-S. 2019. Protective role of freshwater and marine rotifer glutathione S-transferase sigma and omega isoforms transformed into heavy metal-exposed Escherichia coli. Environ Sci Technol. 53(13):7840–7850.
- Michaloudi E, Papakostas S, Stamou G, Neděla V, Tihlaříková E, Zhang W, Declerck S. 2018. Reverse taxonomy applied to the Brachionus calyciflorus cryptic species complex: Morphometric analysis confirms species delimitations revealed by molecular phylogenetic analysis and allows the (re)description of four species. PLoS One. 13(9):e0203168.
- Mills S, Alcántara-Rodríguez JA, Ciros-Pérez J, Gómez A, Hagiwara A, Hinson Galindo K, Jersabek CD, Malekzadeh-Viayeh R, Leasi F, Lee J-S, et al. 2017. Fifteen species in one: deciphering the Brachionus plicatilis species complex (Rotifera, Monogononta) through DNA taxonomy. Hydrobiologia. 796(1):39–58.
- Nie ZJ, Gu RB, Du FK, Shao NL, Xu P, Xu GC. 2016. Monogonont rotifer, Brachionus calyciflorus, possesses exceptionally large, fragmented mitogenome. PLoS One. 11(12):e0168263
- Nys C, Janssen CR, De Schamphelaere KA. 2016. Development and validation of a chronic Pb bioavailability model for the freshwater rotifer Brachionus calyciflorus. Environ Toxicol Chem. 35(12):2977–2986.
- Papakostas S, Michaloudi E, Proios K, Brehm M, Verhage L, Rota J, Peña C, Stamou G, Pritchard VL, Fontaneto D, Declerck SA. 2016. Integrative taxonomy recognizes evolutionary units despite widespread mitonuclear discordance: Evidence from a rotifer cryptic species complex. Syst Biol. 65(3):508–524.
- Paraskevopoulou S, Tiedemann R, Weithoff G. 2018. Differential response to heat stress among evolutionary lineages of an aquatic invertebrate species complex. Biol Lett. 14(11):20180498.
- Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. 2008. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 25(6):1129–1137.
- Xiang XL, Xi YL, Wen XL, Zhang G, Wang JX, Hu K. 2011. Genetic differentiation and phylogeographical structure of the Brachionus calyciflorus complex in eastern China. Mol Ecol. 20(14):3027–3044.
- Zhang G, Xi YL, Xue YH, Xiang XL, Wen XL. 2015. Coal fly ash effluent affects the distributions of Brachionus calyciflorus sibling species. Ecotoxicol Environ Saf. 112:60–67.
