Abstract
The phylogenetic relationships of dragonflies have received great attention all the time. For a better understanding the phylogenies among odonate insects, the paper presented the complete mitochondrial genome of Acisoma panorpoides based on next generation sequencing data of total genomic DNA. The total length comprised 15,249 bp and the 37 genes (2 rRNA genes, 13 protein coding genes and 22 tRNA genes). Gene content and gene arrangement were identical to other odonate mitogenomes. Phylogenetic analyses using the whole sequences of the mitochondrial genome placed A. panorpoides as a sister species to Hydrobasileus croceus in Libellulidae.
The Odonata is consisted of three suborders (Zygoptera, Anisozygoptera and Anisoptera) with approximately 6500 described species. Odonate insects play a key role in the evolution of winged insects and as environmental indicators (Catling Citation2005). The phylogenetic relationships among families of Odonata (Dijkstra et al. Citation2014; Carle et al. Citation2015) have traditionally been controversial. In this paper, we sequenced and annotated the complete mitogenome sequences of Acisoma panorpoides Rambur, 1842 (Odonata: Libellulidae) which will provide the help in understanding the diversity and phylogeny of dragonflies in the future.
The adults of Acisoma panorpoides were collected from Nanchang city of Jiangxi province (28°66′87″N, 115°97′93″E), China, in July 2018. The specimen (ODA-1) was preserved in the herbarium of Jiangxi Normal University, Nanchang, China. The total genomic DNA was extracted from the flight muscles of a single individual using the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s protocol. The whole mitochondrial genome sequencing was performed in Shanghai Majorbio Bio-Pharm Technology Co., Ltd (Shanghai, China). The mitochondrial genome was assembled using SOAPdenovo v2.04 (http://soap.genomics.org.cn/) and annotated via available mitochondrial genomes of other odonate species (Lorenzo-Carballa et al. Citation2014; Yu et al. Citation2014; Feindt et al. Citation2016; Herzog et al. Citation2016). The sequences of the other 18 odonate mitochondrial genomes and Parafronurus youi were downloaded from GenBank and were used to construct the minimum evolutionary tree (10000 replicates) using MEGA X (Kumar et al. Citation2018).
The complete mitochondrial genome sequence of A. panorpoides (GenBank accession no. MN046207) was a typical circular DNA molecule with the length of 15,249 bp. Acisoma panorpoides encoded an A + T-rich control region and a typical set of 37 mitochondrial genes, including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes. The arrangement and orientation of the mitochondrial genes are identical to the other odonate mitochondrial genomes (Simon and Hadrys Citation2013; Wang et al. Citation2015; Feindt et al. Citation2016). The base composition of the mitochondrial genome is A: 34.6%, T: 39.8%, C: 11.1% and G: 14.5%, with an A + T content of 74.4%. All protein coding genes employed the standard start codon for invertebrate mitochondrial genome: two use TTG (cox1 and nad4L); three use ATT (atp8, nad5, and nad6); three use ATA (nad1, nad2, and nad3) and five genes start ATG (atp6, cox3, nad4, cob and cox2). The standard stop codon TAA was used 12 times (nad2, cox1, cox2, cox3, atp8, atp6, nad4L, nad6, nad5, cob, nad1, and nad2), except TAG was used only once by nad3. The length of 22 tRNA ranged from 64 bp to 71 bp and all tRNA genes have the typical cloverleaf structure except for trnS1.
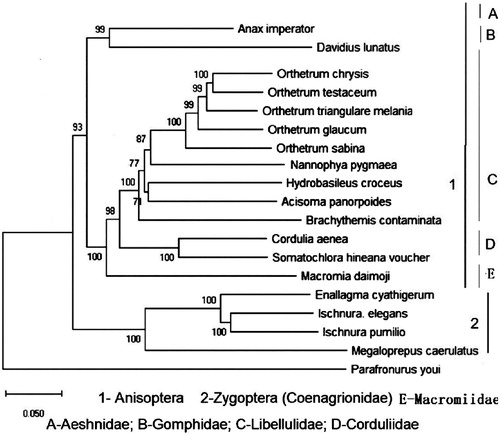
In the mitogenome of A. panorpoides, there were six non-coding intergenic regions including S5 (nad1/trnL1) which often lack in Zygopterans (Herzog et al. Citation2016; Kim et al. Citation2018). The phylogenetic tree supported A. panorpoides as a sister species to Hydrobasileus croceus (). To better understand the odonate phylogeny on multiple taxonomic levels better, it is necessary to increase gradually dragonfly taxon sampling and sequencing.
Figure 1. Mitochondrial phylogenetic relationships of 18 Odonata species obtained using complete sequences. GenBank accession numbers are as follows: Anax imperator (KX161841); Davidius. lunatus (EU591677); Orthetrum chrysis (KU361233); Orthetrum testaceum (KU361235); Orthetrum triangulare melania (AB126005.1); Orthetrum glaucum (KU361232); Orthetrum Sabina (KU361234); Nannophya. pygmaea (KY402222); Hydrobasileus croceus (KM244659); Brachythemis contaminate (KM658172); Cordullia aenea (JX963627); Somatochlora hineana voucher (MG594801.1); Macromia daimoji (MF990748); Enallagma cyathigerum (MF716899); Ischnura elegans (KU958378); Ischnura pumilio (KC878732); Megaloprepus caerulatus (KU958377); Parafronurus youi (EU349015.1).
Geolocation information
The species were collected in Jiangxi Normal University (28°66′87″N, 115°97′93″E), Nanchang city of Jiangxi province, China. The species were preserved in the herbarium of College of Life Science, Jiangxi Normal University, Nanchang, China.
Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- Carle FL, Kjer KM, May ML. 2015. A molecular phylogeny and classification of Anisoptera (Odonata). Arthropod Syst Phylog. 73:281–301.
- Catling PM. 2005. A potential for the use of dragonfly (Odonata) diversity as a bioindicator of the efficiency of sewage lagoons. Can Field Nat. 119:233–236.
- Dijkstra KDB, Kalkman VJ, Dow RA, Stokvis FR, Van Tol J. 2014. Redefining the damselfly families: a comprehensive molecular phylogeny of Zygoptera (Odonata). Syst Entomol. 39:68–96.
- Feindt W, Osigus H-J, Herzog R, Mason CE, Hadrys H. 2016. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondr DNA B. 1:497–499.
- Herzog R, Osigus HJ, Feindt W, Schierwater B, Hadrys H. 2016. The complete mitochondrial genome of the emperor dragonfly Anax imperator LEACH, 1815 (Odonata: Aeshnidae) via NGS sequencing. Mitochondr DNA B. 1:783–786.
- Kim MJ, Jeong SY, Park NS, Wang AR, An J, Kim I. 2018. Complete mitochondrial genome sequence of Macromia daimoji Okumura, 1949 (Odonata: Macromiidae). Mitochondr DNA B. 3:365–367.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lorenzo-Carballa MO, Thompson DJ, Cordero-Rivera A, Watts PC. 2014. Next generation sequencing yields the complete mitochondrial genome of the scarce blue-tailed damselfly, Ischnura pumilio. Mitochondr DNA. 25:247–248.
- Simon S, Hadrys H. 2013. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol Phylogenet Evol. 69:393–403.
- Wang JF, Chen MY, Chaw SM, Morii Y, Yoshimura M, Sota T, Lin CP. 2015. Complete mitochondrial genome of an enigmatic dragonfly, Epiophlebia superstes (Odonata, Epiophlebiidae). Mitochondr DNA. 26:718–719.
- Yu P, Cheng X, Ma Y, Yu D, Zhang J. 2014. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondr DNA A. 27:2271–2273.
