Abstract
Ruta graveolens L. is a perennial plant belonging to the family Rutaceae and has been used for traditional medicines for a long time. In this study, the complete chloroplast (cp) genome sequence of R. graveolens was first reported and characterized. The cp genome is 157,434 bp in length and contains a pair of inverted repeats (IRs, 26,868 bp) separated by a large (85,387 bp) and small (18,311 bp) single-copy regions. A total of 132 genes were predicted, including 87 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The phylogenetic analysis suggested that R. graveolens is more closely related to the monophyletic subfam. Aurantioideae.
Ruta graveolens L. is a perennial herb (Rutoideae, Rutaceae), which is native to the Mediterranean region of southern Europe and northern Africa and now cultivated in many countries of the world (Miguel Citation2003). Since ancient times, R. graveolens has been used in traditional medicines for the relief of pain, eye problems, rheumatism and dermatitis. Recently, R. graveolens has shown various pharmacological activities, including contraceptive, hypotensive, antimicrobial, analgesic, anti-inflammatory, antipyretic, antidiabetic and insecticidal activities (Miguel Citation2003; Salvo et al. Citation2010; Asgarpanah Citation2012; Kannan and Babu Citation2012; Parray et al. Citation2012; Malik et al. Citation2016). At now, more than 120 natural compounds mainly including acridone alkaloids, coumarins, essential oils, flavonoides, and fluoroquinolones have been found in the roots and aerial parts (Asgarpanah Citation2012; Kannan and Babu Citation2012; Parray et al. Citation2012; Malik et al. Citation2016). To provide genomic resources for investigating the evolution of R. graveolens, the complete chloroplast (cp) genome of this species was analyzed from high-throughput Illumina sequencing reads.
The fresh leaves of R. graveolens were collected from Kunming Institute of Botany, CAS (N25°08′21″, E102°44′30″, 1,950 m) and the specimen (lpssy0212) was deposited at the herbarium of the Liupanshui Normal University (LPSNU). The genomic DNA was extracted and sequenced as previously described (Zhang et al. Citation2019). Approximately 2 Gb raw data were generated with pair-end 150 bp read length on the Illumina HiSeq 4000 Platform. The cp genome of R. graveolens was de novo assembled using the GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle) and all genes were annotated using Dual Organellar Genome Annotator (DOGMA) (Wyman et al. Citation2004). The complete cp genome sequence of R. graveolens was deposited in GenBank database (accession no. MN326012).
The complete R. graveolens cp genome is 157,434 bp in length, including a large single copy (LSC) region of 85,387 bp, a small single copy (SSC) region of 18,311 bp, and a pair of inverted repeats (IRs) of 26,868 bp each. The cp genome shows the GC content of 37.2% and contains 132 genes, including 87 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. Of them, 15 distinct (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) contain one intron and three genes (clpP, rps12 and ycf3) have two introns.
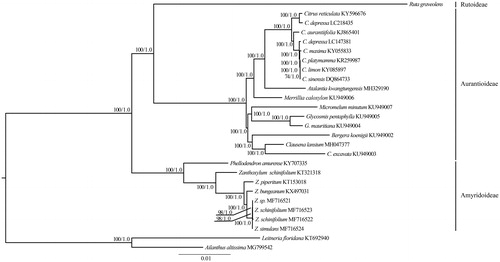
The Rutaceae family comprises 150–170 genera and approximately 2,040 species and includes four subfamilies: Aurantioideae, Cneoroideae, Rutoideae, and Amyridoideae (Morton and Telmer Citation2014). The cp genomes of R. graveolens and previously published species of Rutaceae were used for phylogenetic analysis. In this study, the cp genomes from 24 representative species from two subfamiles (Aurantioideae and Amyridoideae) were downloaded and their GenBank accession numbers are provided in . Two species (Leitneria floridana and Ailanthus altissima) from Simaroubaceae were used as the outgroups. shows that there is very strong support for a monophyletic subfam. Aurantioideae, which is consistent with the previous results based on the cpDNA and nuclear DNA sequences (Groppo et al. Citation2008; Bayer et al. Citation2009; Morton and Telmer Citation2014; Appelhans et al. Citation2016). Ruta graveolens (Rutoideae) is sister to Aurantioideae, which are strongly supported as sister to Phellodendron and Zanthoxylum (Amyridoideae) ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Appelhans MS, Krohm S, Manafzadeh S, Wen J. 2016. Phylogenetic placement of Psilopeganum, a rare monotypic genus of Rutaceae (the citrus family) endemic to China. J Sytem Evol. 54:535–544.
- Asgarpanah J. 2012. Phytochemistry and pharmacological properties of Ruta graveolens L. J Med Plants Res. 6:3942–3949.
- Bayer RJ, Mabberley DJ, Morton C, Miller CH, Sharma IK, Pfeil BE, Rich S, Hitchcock R, Sykes S. 2009. A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. Am J Bot. 96:668–685.
- Groppo M, Pirani JR, Salatino MLF, Blanco SR, Kallunki JA. 2008. Phylogeny of Rutaceae based on two noncoding regions from cpDNA. Am J Bot. 95:985–1005.
- Kannan R, Babu U. 2012. Identity and pharmacognosy of Ruta graveolens Linn. Ancient Sci Life. 32:16–19.
- Malik S, Moraes DFC, do Amaral F, Ribeiro M. 2016. Ruta graveolens: phytochemistry, pharmacology, and biotechnology. In: Jha S, editor. Transgenesis and secondary metabolism. reference series in phytochemistry. Cham: Springer; p. 177–204.
- Miguel ES. 2003. Rue (Ruta L., Rutaceae) in traditional Spain: frequency and distribution of its medicinal and symbolic applications. Econ Bot. 57:231–244.
- Morton CM, Telmer C. 2014. New subfamily classification for the Rutaceae. Ann Mo Bot Gard. 99:620–641.
- Parray SA, Bhat J, Ahmad G, Jahan N. 2012. Ruta graveolens: from traditional system of medicine to modern pharmacology: an overview. Am J PharmTech Res. 2:239–252.
- Salvo G, Ho SYW, Rosenbaum G, Ree R, Conti E. 2010. Tracing the temporal and spatial origins of island endemics in the Mediterranean region: a case study from the citrus family (Ruta L., Rutaceae). Syst Biol. 59:705–722.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang SD, Zhang C, Ling LZ. 2019. The complete chloroplast genome of Rosa berberifolia. Mitochondrial DNA B. 4:1741–1742.