Abstract
To understand genetic features of new candidate cultivar, Dae Ryun of A. distichum, chloroplast genome was sequenced. Its length is 156,019 bp and has four subregions: 86,783 bp of large single-copy (LSC) and 17,828 bp of small single-copy (SSC) regions are separated by 25,704 bp of inverted repeat (IR) regions including 133 genes (87 protein-coding genes, 8 rRNAs, and 37 tRNAs). Overall GC content is 35.8%. Intraspecies sequence variations among four A. distichum chloroplast genomes present various numbers. Phylogenetic trees show that A. distichum is clustered with 12 Forsythia species.
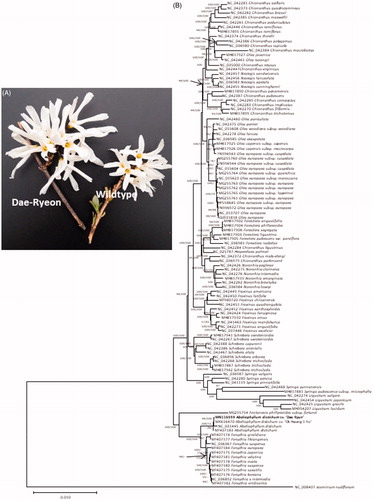
Abeliophyllum distichum Nakai (Oleaceae) is a monotypic genus endemic to Korea (Chung Citation1999). Nine natural habitats have been found (Lee et al. Citation2014). Abeliophyllum distichum displays various phenotypes of colours and size of flowers (Lee Citation1976) and also shows continuous phenotypic variations inside habitat (Kim Citation2001). Ok-Hwang-1-ho became first official cultivar of A. distichum and its chloroplast genome was completed presenting enough number of variations (Park, Kim, Xi, Jang, et al. Citation2019). To unravel relationship between phenotypic and genetic variations of A. distichum with three available chloroplasts (Kim et al. Citation2016; Ha et al. Citation2018; Park, Kim, Xi, Jang, et al. Citation2019), chloroplast of new candidate cultivar, named as Dae Ryun, of which flower is larger than wildtype () was sequenced.
Figure 1. (A) Picture of flowers of Dae Ryun and wildtype of A. distichum. (B) Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 108 Oleaceae chloroplast genomes: Abeliophyllum distichum (MN116559, MN116559, NC_031445, and MF407183), Chionanthus axillaris (NC_042281), Chionanthus quadristamineus (NC_042373), Chionanthus brassii (NC_042282), Chionanthus maxwellii (NC_042385), Chionanthus pedunculatus (NC_042261), Chionanthus ramiflorus (NC_042446 and MH817895), Chionanthus thorelii (NC_042374), Chionanthus polygamus (NC_042386), Chionanthus rupicola (NC_036980), Chionanthus macrobotrys (NC_042384), Olea javanica (MH817927), Olea tsoongii (NC_042461), Chionanthus retusus (NC_035000), Chionanthus virginicus (NC_042447), Nestegis sandwicensis (NC_042457), Nestegis lanceolata (NC_042456), Nestegis apetala (NC_036983), Nestegis cunninghamii (NC_042455), Chionanthus panamensis (MH817890), Chionanthus pubescens (NC_042387), Chionanthus compactus (NC_042269), Chionanthus implicatus (NC_042283), Chionanthus filiformis (NC_042270), Chionanthus trichotomus (MH817899), Olea paniculata (NC_042460), Olea perrieri (NC_042375), Olea woodiana subsp. woodiana (NC_015608), Olea lancea (NC_042278), Olea exasperata (NC_036985), Olea capensis subsp. capensis (MH817925), Olea capensis subsp. macrocarpa (MH817926), Olea europaea subsp. cuspidata (FN996943, MG255760, FN996944, and NC_015604), Olea europaea subsp. guanchica (MG255764), Olea europaea subsp. maroccana (NC_015623), Olea europaea subsp. europaea (MG255763, MG255762, MG255761, HF558645 and FN996972), Olea europaea subsp. laperrinei (MG255765), Olea europaea (NC_013707, GU931818), Forestiera angustifolia (MH817902), Forestiera phillyreoides (MH817904), Forestiera segregata (MH817906), Forestiera ligustrina (MH817903), Forestiera pubescens var. parviflora (MH817905), Forestiera isabelae (NC_036981), Chionanthus ligustrinus (NC_042284), Hesperelaea palmeri (NC_025787), Chionanthus mala-elengi (NC_042372), Chionanthus parkinsonii (NC_036979), Noronhia peglerae (NC_042426), Noronhia clarinerva (NC_042275), Noronhia intermedia (NC_042276), Noronhia emarginata (MH817919), Noronhia brevituba (NC_042262), Noronhia lowryi (NC_036984), Fraxinus americana (NC_042449), Fraxinus latifolia (NC_042450), Fraxinus chiisanensis (MF980720), Fraxinus quadrangulata (NC_042451), Fraxinus xanthoxyloides (NC_042452), Fraxinus lanuginosa (NC_042424), Fraxinus ornus (MH817910), Fraxinus mandshurica (NC_041463), Fraxinus angustifolia (NC_042271), Fraxinus excelsior (NC_037446), Schrebera swietenioides (MH817941 and NC_042267), Schrebera capuronii (NC_042388), Schrebera orientalis (NC_042266), Schrebera alata (NC_042467), Schrebera arborea (NC_036986), Schrebera trichoclada (NC_042268, MH817867 and MH817942), Syringa vulgaris (NC_036987), Syringa persica (NC_042280), Syringa pinnatifolia (NC_041119), Syringa yunnanensis (NC_042468), Syringa pubescens subsp. microphylla (MH817881), Ligustrum vulgare (NC_042274), Ligustrum japonicum (NC_042454), Ligustrum gracile (NC_042425), Ligustrum lucidum (MH394207), Fontanesia phillyreoides subsp. fortunei (MG255754), Forsythia giraldiana (MF407174), Forsythia likiangensis (MF407177), Forsythia suspensa (NC_036367, MF407180), Forsythia europaea (MF407184), Forsythia japonica (MF407175), Forsythia velutina (MF407181), Forsythia ovata (MF407178), Forsythia saxatilis (MF407179), Forsythia koreana (MF407176), Forsythia x intermedia (NC_036982), Forsythia viridissima (MF407182), and Jasminum nudiflorum (NC_008407) as an outgroup. Phylogenetic tree was drawn based on neighbor joining tree. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.
Total DNA of Dae-Ryun collected in GoesanBunjae-Nongwon (Goesan-gun, Chungbuk Province, Korea, Voucher in InfoBoss Cyber Herbarium (IN); IB-01025) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome was sequenced using HiSeqX at Macrogen Inc., Korea, and de novo assembly and sequence confirmation were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation from A. distichum (MK616470; Park, Kim, Xi, Jang, et al. Citation2019).
Chloroplast genome of A. distichum (Genbank accession is MN116559) is 156,019 bp (GC ratio is 37.8%) and has four subregions: 86,783 bp of large single-copy (35.8%) and 17,828 bp of small single-copy (31.9%) regions are separated by 25,704 bp of inverted repeat (IR; 43.2%). It contains 133 genes (87 protein-coding genes, 8 rRNAs, and 37 tRNAs); 18 genes (7 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions.
Based on alignments with three A. distichum chloroplast genomes, 9 single nucleotide polymorphisms (SNPs) and 11 insertions and deletions (INDELs) were found between DaeRyun and Ok-Hwang-1-ho or isolate (MF407183), which is higher than those of Coffea arabica (Park, Kim, Xi, Heo Citation2019; Park, Kim, Xi, Nho, et al. Citation2019; Park, Xi, et al. Citation2019), Marchantia polymorpha (Kwon et al. Citation2019), Camellia japonica (Park, Kim, Xi, Oh, et al. Citation2019), Dysphania pumilio (Park and Kim Citation2019), and Cucumis melo (Zhu et al. Citation2016). While, there are 102 SNPs and 63 INDELs between Dae Ryun and another isolate (NC_031445), similar to those of Pseudostellaria palibiniana (Kim et al. Citation2019), Illicium anisatum (Park, Kim, Xi Citation2019), Duchesnea chrysantha (Park, Kim, Lee Citation2019), and Dioscorea polystachya (Cao et al. Citation2018).
One hundred and eight Oleaceae chloroplast genomes including four A. distichum chloroplasts were used for constructing bootstrapped neighbor joining and maximum likelihood trees using MEGA X (Kumar et al. Citation2018) and IQ-TREE 1.6.6 (Nguyen et al. Citation2015), respectively, after aligning whole chloroplast genomes with modifying directions of subregions by MAFFT 7.388 (Katoh and Standley Citation2013). Trees show that four A. distichum formed independent clade from twelve Forsythia species (.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cao J, Jiang D, Zhao Z, Yuan S, Zhang Y, Zhang T, Zhong W, Yuan Q, Huang L. 2018. Development of chloroplast genomic resources in Chinese Yam (Dioscorea polystachya). BioMed Res Int. 2018: 6293847.
- Chung MG. 1999. Allozyme diversity in the endangered shrub Abeliophyllum distichum (Oleaceae): a monotypic Korean genus. Int J Plant Sci. 160:553–559.
- Ha Y-H, Kim C, Choi K, Kim J-H. 2018. Molecular phylogeny and dating of Forsythieae (Oleaceae) provide insight into the Miocene history of Eurasian temperate shrubs. Front Plant Sci. 9:99.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim DK. 2001. A phylogenetic study of tribe Forsythieae (Oleaceae) based on randomly amplified polymorphic DNA analysis. Daejeon (Korea): Daejeon University.
- Kim H-W, Lee H-L, Lee D-K, Kim K-J. 2016. Complete plastid genome sequences of Abeliophyllum distichum nakai (oleaceae), a Korea endemic genus. Mitochondrial DNA Part B. 1:596–598.
- Kim Y, Heo K-I, Park J. 2019. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): intraspecies variations based on geographical distribution. Mitochondrial DNA Part B. 4:1310–1311.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Kwon W, Kim Y, Park J. 2019. The complete mitochondrial genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: inverted repeats on mitochondrial genome between Korean and Japanese isolates. Mitochondrial DNA Part B. 4:769–770.
- Lee H-Y, Kim T-G, Oh C-H. 2014. Recently augmented natural habitat of Abeliophyllum distichum Nakai in Yeoju-si, Gyunggi-do. Korean J Environ Ecol. 28:62–70.
- Lee TB. 1976. Vascular plants and their uses in Korea. Bull Kwanak Arb. 1:1–137.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:1303.3997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Park J, Kim Y. 2019. The second complete chloroplast genome of Dysphania pumilio (R. Br.) mosyakin & clemants (Amranthaceae): intraspecies variation of invasive weeds. Mitochondrial DNA Part B. 4:1428–1429.
- Park J, Kim Y, Lee K. 2019. The complete chloroplast genome of Korean mock strawberry, Duchesnea chrysantha (Zoll. & Moritzi) Miq.(Rosoideae). Mitochondrial DNA Part B. 4:864–865.
- Park J, Kim Y, Xi H. 2019. The complete chloroplast genome of aniseed tree, Illicium anisatum L. (Schisandraceae). Mitochondrial DNA Part B. 4:1023–1024.
- Park J, Kim Y, Xi H, Heo K-I. 2019. The complete chloroplast genome of ornamental coffee tree, Coffea arabica L. (Rubiaceae). Mitochondrial DNA Part B. 4:1059–1060.
- Park J, Kim Y, Xi H, Jang T, Park J-H. 2019. The complete chloroplast genome of Abeliophyllum distichum Nakai (Oleaceae), cultivar Ok Hwang 1ho: insights of cultivar specific variations of A. distichum. Mitochondrial DNA Part B. 4:1640–1642.
- Park J, Kim Y, Xi H, Nho M, Woo J, Seo Y. 2019. The complete chloroplast genome of high production individual tree of Coffea arabica L. (Rubiaceae). Mitochondrial DNA Part B. 4:1541–1542.
- Park J, Kim Y, Xi H, Oh Y-J, Hahm KM, Ko J. 2019. The complete chloroplast genome of common camellia tree, Camellia japonica L. (Theaceae), adapted to cold environment in Korea. Mitochondrial DNA Part B. 4:1038–1040.
- Park J, Xi H, Kim Y, Heo K-I, Nho M, Woo J, Seo Y, Yang JH. 2019. The complete chloroplast genome of cold hardiness individual of Coffea arabica L. (Rubiaceae). Mitochondrial DNA Part B. 4:1083–1084.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
- Zhu Q, Gao P, Liu S, Amanullah S, Luan F. 2016. Comparative analysis of single nucleotide polymorphisms in the nuclear, chloroplast, and mitochondrial genomes in identification of phylogenetic association among seven melon (Cucumis melo L.) cultivars. Breed Sci. 66:711–719.
