Abstract
The complete mitochondrial genome of Ostorhinchus fleurieu was first determined, which was 16,521 bp in length, containing 13 protein-coding genes, two rRNA genes, 22 tRNA genes, a putative control region and one origin of replication on the light-strand. The overall base composition included C(29.2%), A(26.7%), T(26.7%) and G(17.4%). Moreover, the 13 PCGs encoded 3800 amino acids in total, twelve of which used the initiation codon ATG except for COI started with GTG. Most of them ended with complete stop codon, whereas three protein-coding genes (COII, ND4 and Cytb) used incomplete stop codon and represented as T. The phylogenetic tree based on the Neighbour Joining method was constructed to provide relationship within Apogoninae, which could be a useful basis for management of this species.
The flower cardinalfish (Ostorhinchus fleurieu) distributes in the Visakhapatnam waters, Middle East coast of India (Randall and Hayashi Citation1990), and it is common in shallow coastal reefs with moderate currents, also in tidal channels of estuaries (Fraser Citation2014). The genus Ostorhinchus is composed of nearly 60 species, however, the genetic and molecular information of Ostorhinchus is limited, plus there are no any complete mitogenomes of Ostorhinchus have been determined (Fraser Citation2014), these factors jointly contributed to the motivation of sequencing this species. To gain its molecular information, herein, we described the complete mitogenome of O. fleurieu and explored the phylogenetic relationship within Apogoninae, contributing to further phylogenetic studies on its related species.
Specimen was collected from Nandu river, Haina province, China (18°29′48″N; 109°56′12″E) and stored in the Molecular Breeding Laboratory of Zhejiang Ocean University with accession number 20150826SP22.
The mitogenome of O. fleurieu was a closed double-stranded circular molecule of 16,521bp (GenBank accession No. MN381712), which contained 13 PCGs, 2 rRNA and 22 tRNA genes, and 2 main non-coding regions; nucleotide composition of the complete mitogenome was biased toward A and T, with a slightly higher content of A + T (53.4%) than G + C; genes were not coded totally by a single, most of them located on the heavy strand while ND6 and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro) were detected on the light strand, these features are identical to those of other vertebrates (Cui et al. Citation2009; Jing et al. Citation2010; Zhu et al. Citation2018a). 13 PCGs encoded 3,800 amino acids in total, all PCGs began with a canonical start codon NTG. For the stop codon, most of PCGs were terminated with the complete codon TAA, TAG or AGA, whereas three PCGs (COII, ND4 and Cytb) located immediately upstream of tRNAs ended with only T, some authors suggested that these truncated stop codons were caused by post-transcriptional polyadenylation (Ojala et al. Citation1981).
The small and large subunit ribosomal RNAs (12S and 16S) were 957 and 1687 bp, respectively. They were located in the typical positions between tRNA-Phe and tRNA-Leu(UUA), separated by tRNA-Val. The origin of light-strand replication was located in a cluster of five tRNA genes (WANCY) as in other vertebrates (Zhu et al. Citation2018b, Citation2018c, Citation2018d), which has the capacity of folding into a stable stem-loop secondary structure; the CR was determined to be 854 bp, by comparing the sequences of the CR with other teleost, three typical domains were noticed, including termination-associated sequences, the central conserved sequence block domain and the conserved sequence block domain, which is identical to that in other teleostean mitogenomes (Ruokonen and Kvist Citation2002).
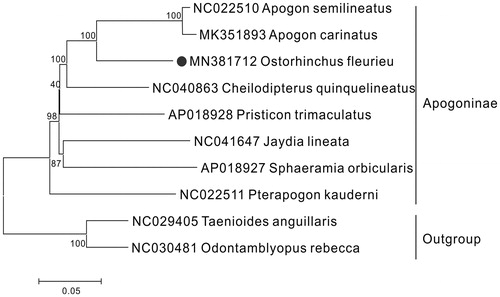
The results of phylogenetic analysis shown that O. fleurieu was closely related to the species of Apogon, with a highly supported value of 100% (). The sequence and full annotation of O. fleurieu would provide more molecular resources about Apogon mitogenomes and lay a foundation for the further research involved with phylogenetic relationship within Apogoninae.
Figure 1. The phylogenetic tree of Ostorhinchus fleurieu and other 7 Apogoninae species was constructed using the Neighbour Joining (NJ) methods based on 12 protein-coding genes encoded by the heavy strand. The bootstrap values are based on 1,000 resamplings and the number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labelled with a black spot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cui Z, Yuan L, Chi PL, Feng Y, Chu KH. 2009. The complete mitochondrial genome of the large yellow croaker, Larimichthys crocea (Perciformes, Sciaenidae): Unusual features of its control region and the phylogenetic position of the Sciaenidae. Gene. 432:33–43.
- Fraser TH. 2014. Foa yamba, a new species of cardinalfish (Percomorpha: Apogonidae: Apogonichthyini) from the tidal region of the Clarence River, Australia and redescriptions of the West Pacific Foa longimana and Foa hyalina. Zootaxa. 3878:167–178.
- Jing H, Zhang D, Hao J, Huang D, Cameron S, Zhu C. 2010. The complete mitochondrial genome of the yellow coaster, Acraea issoria (Lepidoptera: Nymphalidae: Heliconiinae: Acraeini): sequence, gene organization and a unique tRNA translocation event. Mol Biol Rep. 37:3431–3438.
- Ojala D, Montoya J, Attardi G. 1981. TRNA punctuation model of RNA processing in human mitochondrial. Nature. 290:470–474.
- Randall JE, Hayashi M. 1990. Apogon selas, a new cardinalfish from the western pacific. Jpn J Ichthyol. 36:399–403.
- Ruokonen M, Kvist L. 2002. Structure and evolution of the avian mitochondrial control region. Mol Phylogenet Evol. 23:422–432.
- Zhu K, Gong L, Jiang L, Liu L, Lü Z, Liu B-J. 2018a. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondr DNA Part B. 3:536–537.
- Zhu K, Li G, Lü Z, Liu L, Jiang L, Liu B. 2018b. The complete mitochondrial genome of Chaetodon octofasciatus (Perciformes: Chaetodontidae) and phylogenetic studies of Percoidea. Mitochondr DNA Part B. 3:531–532.
- Zhu K, Lü Z, Liu B, Gong L, Jiang L, Liu L. 2018c. The complete mitochondrial genome of Hyporhamphus quoyi (Beloniformes; Hemiramphidae). Mitochondr DNA Part B. 3:1235–1236.
- Zhu K, Lü Z, Liu L, Gong L, Liu B. 2018d. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondr DNA Part B. 3:301–302.
