Abstract
The Paracheirodon axelrodi is an important freshwater fish in the Amazon basin of South America, more expensive than Paracheirodon innesi. Here, we describe the complete 17,100 base pair (bp) mitochondrial genome of Paracheirodon axelrodi mohavensis. The mitogenome has a nucleotide base composition of A (29.62%), T (29.41%), G (15.39%), and C (25.62%), and encodes 13 protein subunits, 22 tRNAs, a 12S rRNA of 952 bp and 16S rRNA of 1665 bp, and a 1433 bp D-loop control region, each located in the conserved mtDNA structure typical for Characidae fishes. All protein-coding genes have initiation codons of ATG, ND1, ND2, CO2, ND3, ND4L, ND5, ND6 and Cytb ended by TAA as a stop codon, ND4 ended by AGA as a stop codon, CO1 ended by AGG as a stop codon, ATP8 and CO3 ended by TAG as a stop codon, ATP6 ended by ATT as a stop codon. This characterised mitogenome may help inform management practices for Paracheirodon axelrodi mohavensis by facilitating future studies on how allopatric populations of this imperilled species are evolving across refuge habitats.
The Paracheirodon axelrodi is a species of the family Characiformes, which are distributed in slow-flowing rivers in Brazil, Colombia, and Venezuela (Mills and Vevers Citation1982; Beheregaray et al. Citation2010). Paracheirodon axelrodi has a certain difficulty in breeding, and the water temperature is stricter, more precious than other Paracheirodon innesi. Based on these characteristics, there were few reports about its basic biology data including genetic information. Identification of complete mitogenome of the Paracheirodon axelrodi would supplement the limited data on molecular level and be reference for systematics.
The specimen was collected from Amazon Basin, South America (15°31′5″S 71°45′55″W) and stored in a refrigerator of −80 °C in Zhejiang Engineering Research Centre for Mariculture and Fishery Enhancement Museum (Accession number: PA180217). Total genomic DNA was extracted from muscle tissue of individual using the phenol–chloroform method (Barnett and Larson Citation2012; Toni et al. Citation2018). The calculation of base composition and phylogenetic construction was conducted by MEGA6.0 software (Kumar et al. Citation1994; Tamura et al. Citation2013). Similar to the typical mitogenome of vertebrates, the mitogenome of Paracheirodon axelrodi is a closed double-stranded circular molecule of 17,100 bp (GenBank accession MH998225), which contains 13 protein-coding genes, 2 ribosomal RNA genes, 22 tRNA genes, and 1 main non-coding regions (Boore Citation1999; Zhu et al. Citation2018a; Zhu et al. Citation2018b). The overall base composition is 29.62%, 29.41%, 25.58%, and 15.39% for A, T, C, and G, respectively. Most mitochondrial genes are encoded on H-strand except for ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu and tRNA-Pro), which are encoded on the L-strand. All of them use the initiation codon ATG, which is quite common in vertebrate mtDNA (Miya et al. Citation2001; Liu et al. Citation2016). Most of them have TAA as the stop codon, whereas CO1 ends with AGG, ATP6 ends with ATT, ND4 ends with AGA, and two protein-coding genes (CO3 and ATP8) ended with TAG. The lengths of 12S rRNA located between tRNAPhe and tRNAVal and 16S rRNA located between tRNAVal and tRNALeu were 952 bp and 1665 bp, respectively.
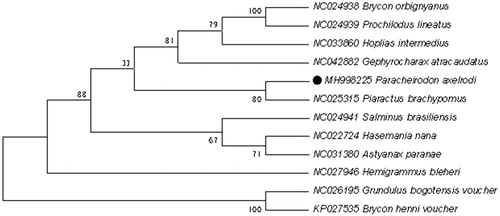
To explore the phylogenetic position of this Paracheirodon axelrodi, a phylogenetic tree was constructed based on the NJ analysis of 12 PCGs encoded by the heavy strand. The results of the present study supports Paracheirodon axelrodi has a closest relationship with Prochilodus lineatus, highly supported by a bootstrap value of 80 (), which are consistent with the results based on morphology and other molecular methods.
Figure 1. Neighbour-joining (NJ) tree of 12 Beloniformes species based on 12 PCGs encoded by the heavy strand. The bootstrap values are based on 1000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labelled with a black spot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol Biol. 840:13–19.
- Beheregaray LB, Möller LM, Schwartz TS, et al. 2010. Microsatellite markers for the cardinal tetra Paracheirodon axelrodi, a commercially important fish from central Amazonia. Mol Ecol Notes. 4:330–332.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Kumar S, Tamura K, Nei M. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci. 10:189–191.
- Li Y, Xue D, Gao T, Liu J. 2016. Genetic diversity and population structure of the roughskin sculpin (Trachidermus fasciatus Heckel) inferred from microsatellite analyses: implicationsfor its conservation and management. Conserv Genet. 17:921–930.
- Mills D, Vevers G. 1982. The Practical Encyclopedia of Freshwater Tropical Aquarium Fishes. Melbourne (Australia): Lansdowne Press.
- Miya M, Kawaguchi A, Nishida M. 2001. A case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18:1993–2009.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Toni LS, Garcia AM, Jeffrey DA, Jiang X, Stauffer BL, Miyamoto SD, Sucharov CC. 2018. Optimization of Phenol-Chloroform RNA extraction. Methodsx. 5:599–608.
- Zhu K, Lu Z, Liu L, Gong L, Liu B. 2018b. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3:301–302.
- Zhu K, Gong L, Jiang L, Liu L, Lü Z, Liu B-J. 2018a. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondrial DNA Part B. 3:536–537.
