Abstract
We report the complete mitochondrial genome sequence of Poecilia formosa. The genome is found to be 16636 bp in length and has a base composition of A (29.59%), G (14.61%), C (28.26%), and T (27.54%). Similar to other Poecilia species, it contains a typically conserved structure including 13 protein-coding genes, 2 rRNA genes, 1 control region (D-loop), and 22tRNA genes. The proportion of coding sequences with a total length of 11,533 bp is 69.33%, which encodes 3837 amino acids. All protein-coding genes started with Met, ND1, CO1, ATP8, ATP6, CO3, ND4L, ND5, ND6 and CytB ended by TAA as a stop codon, ND2 and ND3 ended by TAG as a stop codon, CO3 and ND4 ended by a single T. The lengths of 12S ribosomal RNA is 948 bp, ranging from 70 bp to 1018 bp, and the lengths of 16S ribosomal RNA is 1674 bp, ranging from 1090 bp to 2764 bp. The length of control region is 879 bp, ranging from 15757 bp to 16636 bp, respectively. The complete mitochondrial genome sequence provided here would be useful for further understanding the evolution of ratite and conservation genetics of Poecilia formosa.
Poecilia formosa, belongs to the family Poecilia, Cyprinodontidae, is a freshwater aquarium fish which distributes in the North America, Texas, and northern Mexico, Nueces River and Grande River (Schlupp et al. Citation2002). Although two complete mitochondrial (mt) genomes belonging to Poecilia formosa have been determined, the Poecilia formosa mt genome sequence has not been reported yet.
Here, we sequenced and characterised the complete mt genome of Poecilia formosa. The specimen was collected from the part of Nueces River between Three Rivers and George West (36°26′11″N, 118°49′24″W). Then it stored in a refrigerator of −80 °C in Zhejiang Engineering Research Centre for Mariculture and Fishery Enhancement Museum(Accession number PF180513). Total genomic DNA was extracted from muscle tissue of individual using the phenol-chloroform method (Barnett and Larson Citation2012). The calculation of base composition and phylogenetic construction were conducted by MEGA6.0 software (Tamura et al. Citation2013). The transfer RNA (tRNA) genes were generated using the programme tRNAs-can-SE (Lowe and Eddy Citation1997). The mitochondrial genome sequence of Poecilia formosa with the annotated genes was deposited in GenBank with the accession number of KT715811.
Similar to the typical mitogenome of vertebrates, the mitogenome of Poecilia formosa is a closed double-stranded circular molecule of 16636 nucleotides (GenBank accession No. KT715811), which contains 13 protein-coding genes, 2 ribosomal RNA genes, 22 tRNA genes and 2 main non-coding regions (Boore Citation1999; Zhu et al., Citation2018a; Zhu et al., Citation2018b). The contents of A, G, T and C are 29.59%, 14.61%, 27.54%, and 28.26%, respectively. Most mitochondrial genes are encoded on H-strand except for ND6 and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu and Pro), which are encoded on the L-strand. The proportion of coding sequences with a total length of 11,533 bp is 69.33%, 13 protein-coding (PCGs) genes encode 3837 amino acids in total. All protein-coding genes started with Met, which is quite common in vertebrate mtDNA (Miya et al. Citation2001; Liu et al. Citation2017). ND1, CO1, ATP8, ATP6, CO3, ND4L, ND5, ND6 and CytB ended by TAA as a stop codon, ND2 and ND3 ended by TAG as a stop codon, CO3 and ND4 ended by a single T. The 12S rRNA and 16S rRNA are 948 and 1674 bp, which are both located in the typical positions between tRNA-Phe and tRNA-Leu (UUA), separated by tRNA-Val. The origin of light-strand replication is located in a cluster of five tRNA genes (WANCY) as in other vertebrates (Petrillo et al. Citation2006), The length of control region is 879 bp, ranging from 15757 bp to 16636 bp.
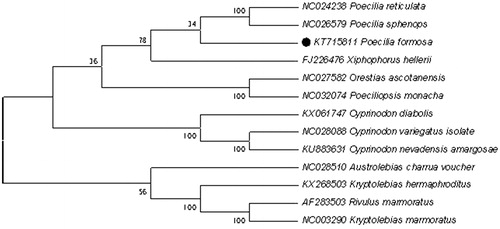
Phylogenetic analysis included mt genome of Poecilia formosa and the other 13 species that are from the order Kryptolebias, Poeciliopsis, Rivulus, Xiphophorus, Poecilia, Orestias, Cyprinodon and Austrolebias, which belong to Cyprinodontiformes. The results of the present study support that Poecilia formosa has a closest relationship with Poecilia reticulata and Poecilia sphenops ().
Figure 1. Phylogenetic tree derived from NJ based on concatenated nucleotide sequences of twelve PCGs. Including Poecilia reticulata, Poecilia sphenops, Poecilia formosa, Poeciliopsis monacha, Xiphophorus hellerii, Orestias ascotanensis, Cyprinodon diabolis, Cyprinodon veriegatus isolate, Cyprinodon nevadensis amargosae, Austrolebias charrua voucher, Kryptolebias hemaphroditus, Rivulus marmoratus,.Kryptolebias marmoratus.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Method Mol Biol. 840:13–19.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Liu J, Ding Q, Gao L. 2017. The complete mitochondrial genome of North Island brown kiwi (Apteryx mantelli). Mitochondrial DNA Part B. Issue 1.
- Lowe T, Eddy S. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Miya M, Kawaguchi A, Nishida M. 2001. A case study for Moderate-Scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18:1993–2009.
- Petrillo M, Silvestro G, Nocera PPD, Boccia A, Paolella G. 2006. Stem-loop structures in prokaryotic genomes. BMC Genomics. 7:170.
- Schlupp I, Parzefall J, Schartl M. 2002. Biogeography of the amazon molly, Poecilia formosa[J]. J Biogeogr. 29:1–6.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhu K, Gong L, Jiang L, Liu L, Lü Z, Liu B-J. 2018a. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondrial DNA Part B. 3:536–537.
- Zhu K, Lü Z, Liu L, Gong L, Liu B. 2018b. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3:301–302.
