Abstract
Complete mitochondrial genome of the characiform fish Hemigrammus bleheri was characterized in the present study. The whole mitogenome was 17,021 bp in size and consisted of 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs genes, a control region, and origin of light-strand replication. The proportion of coding sequences with a total length of 11,415 bp is 67.06%, which encodes 3805 amino acids. Similar to other Hemigrammus species, the base composition of H. bleheri was 29.30% for A, 25.26% for C, 16.36% for G, and 29.08% for T. All PCGs started with Met. ND1, ND3, ND4L, ND6, and CytB ended with TAA as the stop codon. ND2, ATP8, and ND5 ended with TAG as a stop codon, CO2, ATP6, CO3, and ND4 ended simply by T, and CO1 ended by a single AGG. The lengths of 12S ribosomal RNA and 16S ribosomal RNA were 924 bp and 1681 bp, respectively. The length of control region (D-loop) was 1308 bp, ranging from 15,714 to 17,021 bp. The complete mitochondrial genome sequence provided here would be helpful in further understanding the evolution of characiformes and conservation genetics of H. bleheri.
Hemigrammus bleheri belongs to the family Characidae and the order Characiformes, This species is mainly distributed in the Rio Negro and Rio Meta basins. However, few reports about its basic biology data including genetic information could be indexed up to the present. In this study, we first determined the complete mitochondrial genome of H. bleheri, which would provide us the basic molecular data for further study on its systematics and conservation biology.
In the present study, specimens of H. bleheri were collected from the Rio Negro basin of Colombia (3°08′00″N, 59°54′30″W) and stored in a refrigerator at −80°C in Zhejiang Engineering Research Centre for Mariculture and Fishery Enhancement Museum (Accession number: HB180620). Total genomic DNA was extracted from muscle of three different individuals using the phenol–chloroform method (Barnett and Larson Citation2012; Meng et al. Citation2019). The calculation of base composition and phylogenetic construction was conducted by MEGA6.0 software (Tamura et al. Citation2013). The transfer RNA (tRNA) genes were generated with the programme tRNAs-can-SE (Lowe and Eddy Citation1997). The mitochondrial genome sequence of H. bleheri with the annotated genes was deposited in GenBank with the accession number of MK263671.
Similar to the typical mitogenome of vertebrates, the mitogenome of H. bleheri is a closed double-stranded circular molecule of 17,021 nucleotides including 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 tRNA genes, and 2 main noncoding regions (Boore Citation1999; Zhu et al. Citation2018). The contents of A, G, T, and C are 28.67%, 15.86%, 24.37%, and 31.10%, respectively. Most mitochondrial genes are encoded on the H-strand except for ND6 and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro), which are encoded on the L-strand. The proportion of coding sequences with a total length of 11,415 bp is 67.42%, 13 PCGs encode 3805 amino acids in total. A-T and G-C contents of mitochondrial genome are 58.17% and 32.73% respectively.
All the PCGs use the initiation codon ATG, which is quite common in vertebrate mtDNA (Miya et al. Citation2001; Liu et al. Citation2017). ND1, ND3, ND4L, ND6, and CytB ended with TAA as a stop codon, ND2, ATP8, and ND5 ended with TAG as a stop codon, CO1 ended with a single AGG, and four incomplete termination codons (T) were found in the other four genes (CO2, ATP6, CO3, and ND4). The lengths of 12S ribosomal RNA and 16S ribosomal RNA are 924 bp and 1681 bp, which are both located in the typical positions between tRNA-Phe and tRNA-Leu (UUA), separated by tRNA-Val (Petrillo et al. Citation2006; Huang et al. Citation2019). The length of control region (D-loop) is 1308 bp, ranging from 15,714 to 17,021 bp.
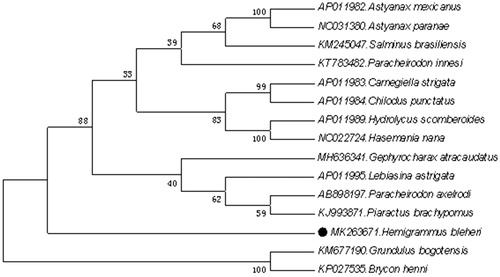
In the neighbour-joining (NJ) tree, the result suggested that H. bleheri was most closely related to Grundulus bogotensis among all the Characiformes species included in the analysis. This result is consistent with conventional morphological taxonomy ().
Figure 1. Neighbour-joining (NJ) tree of 15 Characiformes species based on 12 PCGs. The bootstrap values are based on 10,000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labelled with a black spot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol Biol. 840:13–19.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Huang Y, Liu B, Zhu K, Zhang J, Jing F, Xia L, Liu Y. 2019. The complete mitochondrial genome of Gephyrocharax atracaudatus (Characiformes, Characidae) and phylogenetic studies of Characiformes. Mitochondrial DNA Part B. 4(1):1901–1902.
- Liu J, Ding Q, Gao L. 2017. The complete mitochondrial genome of North Island brown kiwi (Apteryx mantelli). Mitochondrial DNA Part B. 2(1):1–2.
- Lowe T, Eddy S. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res.
- Meng F, Huang Y, Liu B, Zhu K, Zhang J, Jing F, Xia L, Liu Y. 2019. The complete mitochondrial genome of Lebiasina astrigata (Characiformes: Lebiasinida) and phylogenetic studies of Characiformes. Mitochondrial DNA Part B. 4(1):579–580.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18(11):1993–2009.
- Petrillo M, Silvestro G, Nocera PPD, Boccia A, Paolella G. 2006. Stem-loop structures in prokaryotic genomes. BMC Genomics. 7(1):170.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Zhu K, Lu Z, Liu L, Gong L, Liu B. 2018. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3(1):301–302.
