Abstract
Narcissus tazetta var. chinensis is an important economic perennial herb in China. In this study, the complete plastid genome was assembled from the next-generation sequencing data. The circular genome is 159,376 bp long, and comprises a pair of inverted repeat regions (28,492 bp for each), a large single-copy region (85,941 bp), and a small single-copy region (16,451 bp). It encodes 133 genes, including 87 protein-coding genes, 38 tRNAs genes, and 8 rRNA genes. The GC content of N. tazetta var. chinensis cp genome is 38%. Phylogenetic analysis indicates that N. tazetta var. chinensis was closely relate to Narcissus porticus. The complete plastome genome will provide a useful genetic resource for the conservation and garden utilization in the future.
Narcissus tazetta L. var. chinensis Roem. known as Chinese Narcissus or bunch-flowered daffodil belongs to Narcissus L. in Amaryllidaceae (Chen et al. Citation2003). It is one of the top ten traditional flowers in China and has great application value in ornamental as well as refining essential oil (Jian et al. Citation2006). The research on its flowering mechanism is also a hotspot topic in recent years (Wang et al. Citation2018). Narcissus tazetta var. chinensis is triploid with complex genetic background (Zhang et al. Citation2018), which limits its breeding and related theoretical research to a certain extent. In this study, the complete plastid genome was assembled and characterised for the first time in order to provide help for phylogenetic and genetic studies.
Fresh leaves of N. tazetta subsp. chinensis was collected from a single individual at the natural population at (Nanji island, Zhejiang, China; 121.093313E, 27.469608 N). The voucher specimen was deposited at the herbarium of Nanjing Forestry University (NF, accession number NF000015) and DNA compounds were stored at −20 °C. The experiment procedure is as reported in Duan et al. (Citation2019). In all, 19.2 Gb Clean Data were obtained, then assembled using NOVOPlasty with Agapanthus coddii F.M. Leight. (NC035971.1) as reference. A total of 159,376 individual reads yielded an average coverage of 1764 X. The resultant genome was annotated by CpGAVAS (Liu et al. Citation2012).
The circular genome (MN432153) of N. tazetta subsp. chinensis was 159,376 bp in size including two inverted repeat regions of 28,492 bp, which is separated by a large single copy region of 85,941 bp and a small single copy region of 16,451 bp. The overall G + C content of N. tazetta subsp. chinensis genome was 38% (A = 30.8%, T = 19.3%, C = 18.7%, G = 31.2%). A total of 133 genes are encoded, including 87 protein-coding genes, 38 tRNAs and eight rRNAs. Most of the genes occurred in a single copy, however 29 protein-coding genes, 11 tRNA genes, and one rRNA genes are duplicated. Gene synteny and arrangement anaylsis revealed that N. tazetta subsp. chinensis maintained consistent positions, directions, and nucleotide sequences and presented high levels of similarity in most of the genes which mean these plastomes are completely syntenic within Narcissus. While, the relatively lower identity regions were recognized: trnK-UUU, matK, atpI, rpl16, petB, petD, ycf1, and ycf2 regions. Non-coding regions exhibited highly divergent regions include: psbA-chlB, psbM-clpP, ycf4-accD, ycf3-psaA, psaC-ccsA, ndhH- psaC, ycf3-psaA, trnG-UUU and petL-psbF.
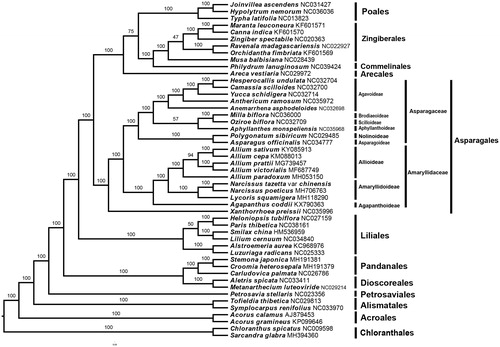
The maximum likelihood phylogeny was reconstructed using RAxML (Stamatakis et al. Citation2008) with 1000 bootstraps under the GTRGAMMAI substitution model in CIPRES (Miller et al. Citation2010). The phylogenetic analysis was conducted based on the concatenate of 69 common protein-coding regions which were extracted from the published plastid genome of 49 monocots species. The result showed that Narcissus tazetta var. chinensis was closely related to Narcissus porticus L. (), furthermore, the phylogeny tree topologically support the set of relationships: [Agapanthaceae [Alliaceae + Amaryllidaceae]] (Givnish et al. Citation2016; Li et al. Citation2016) which is named “Core” (higher) Asparagales with high-resolution value.
Acknowledgments
The authors are grateful to Mingzhi Li at Biodata Biotechnologies Co. Ltd., Hefei for providing technical assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen J, Chen JP, Langeveld SA, Derks AFLM, Adams MJ. 2003. Molecular Characterization of Carla‐ and Potyviruses from Narcissus in China. J Phytopathol. 151(1):26–29.
- Duan Y, Li Y, Zhang C, Wang X, Li M. 2019. The complete chloroplast genome of sweet olive (Osmanthus fragrans Lour.). Mitochondrial DNA B. 4(1):1063–1064.
- Givnish TJ, Zuluaga A, Marques I, Lam VKY, Gomez MS, Iles WJD, Ames M, Spalink D, Moeller JR, Briggs BG, et al. 2016. Phylogenomics and historical biogeography of the monocot order Liliales: out of Australia and through Antarctica. Cladistics. 32(6):581–605.
- Jian WU, Chen LJ, Li GU, Wang YY, Tian HQ. 2006. Embryological studies on Narcissus tazetta var. chinensis. Front Biol China. 1:18–22.
- Li QQ, Zhou SD, Huang DQ, He XJ, Wei XQ. 2016. Molecular phylogeny, divergence time estimates and historical biogeography within one of the world’s largest monocot genera. AoB Plants. 8:pii: plw041.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environment. New Orleans: IEEE; p. 1–8.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771.
- Wang G, Yang B, Wu J, Luo P, Anwar M, Allan AC, Lin-Wang K, Espley RV, Zeng L. 2018. Identification of genes involved in flavonoid biosynthesis of Chinese Narcissus (Narcissus tazetta L. var. chinensis). Plant Mol Biol Rep. 36(5–6):812–821.
- Zhang C, Wentao YU, Min LI, Zhang H, Xing D, Chen Q, Shen J. 2018. Identification of DNA barcodes in Narcissus. J Fujian Agric For Univ. 47:452–460.