Abstract
In the present study, the complete mitochondrial genome of H. grayi was determined and annotated. The circular mitogenome is 16,959 bp in length and contains 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and a control region. Most of the PCGs start with ATG, but CO1 begins with a GTG start codon. Phylogenetic analysis revealed that H. grayi was strictly related to network pipefish Corythoichthys flavofasciatus with 100% bootstrap support value. This work provides basic molecular information that would be useful for further investigation on conservation genetics and evolutionary relationships of H. grayi.
The Gray’s pipefish Halicampus grayi (Kaup, 1856) is a widespread Indo-Pacific species that inhabits muddy sand, estuaries, and coral reefs with a maximum depth of 100 m (Dawson Citation1982). The species is one of the most important resources for syngnathids which have been traded heavily throughout the region for traditional medicines and the aquarium trade (Vincent et al. Citation2011). Due to the strong market demand of dried pipefish, the wild-caught specimens of H. grayi were often traded as main substitutions for Chinese medicinal pipefishes (Gao et al. Citation2018). Furthermore, partial mitochondrial DNA barcoding markers of 16S and COI genes failed to distinguish certain pipefishes as distinct species (Garcia et al. Citation2019). The Complete mitochondrial genome of Gray’s pipefish could provide the basic molecular data and contribute to the development of molecular identification and further conservation strategy (Chen et al. Citation2018; Zhu et al. Citation2019).
In this study, we sequenced the complete mitochondrial genome of H. grayi and investigated the phylogenetic relationship of Genus Halicampus among Syngnathidae species. The specimen of H. grayi was collected from Bozhou herbal medicine market of Anhui Province and identified according to the typical morphological characteristics including the relative short snout and large eyes. The sample of H. grayi GSH-06 was deposited in the collection centre of Zhejiang Chinese Medical University. The entire mtDNA of this pipefish species was sequenced by universal primers according to the previous researches (Ge et al. Citation2018; Lai et al. Citation2019). The complete mtDNA sequence of H. grayi, with the annotated genes, was submitted to GenBank with the accession number of MN064721.
The complete mitochondrial genome of H. grayi was 16,959 bp in length with the overall base compositions of 30.03, 29.25, 24.54, and 16.17% for A, T, C, and G, respectively. The high A + T content bias (59.28%) was basically consistent with those of other syngnathid fishes (Wang et al. Citation2019). The mitogenome of H. grayi, in total, contained 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region. Among these genes, all PCGs genes have a typical ATG initiation codon except CO1 which starts with GTG. The complete stop codon TAA was found in seven PCGs (CO2, ATP8, ATP6, CO3, ND4L, ND5, and ND6), and the remaining six genes terminated with incomplete TA or T. The 22 tRNA genes ranged from 67 to 74 bp in length with the typical clover-leaf structure except tRNA-Ser (AGC) which lacks the dihydrouridine arm. The control region was 1126 bp in length which was significantly longer than that in other Syngnathidae mitogenomes (Fang et al. Citation2018; Sun et al. Citation2019).
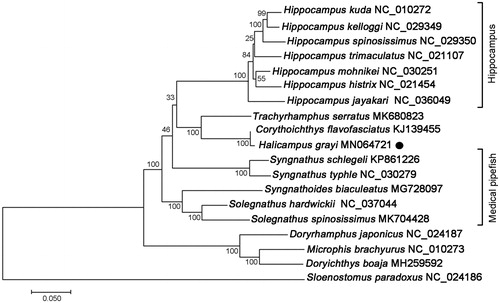
The phylogenetic tree of H. grayi and other Syngnathidae species were constructed by maximum-likelihood (ML) method based on the complete mitochondrial protein coding sequences. As showing in , H. grayi and Corythoichthys flavofasciatus clustered together with high statistical support, indicating a close genetic relationship. Furthermore, the Gray’s pipefish H. grayi shared obviously high nucleotide sequence identity (99.45%) with the network pipefish C. flavofasciatus, suggesting further taxonomic revision among the two syngnathid species (Zhang et al. Citation2015). The genera Halicampus, Corythoichthys, and Trachyrhamphus clustered into a monophyletic group with high bootstrap values, which is sister to the clade clustered by Hippocampus species. These results suggested that fishes in the three genera could be developed as potential alternatives to seahorse species. This study provides basic molecular data of H. grayi, which was important for the development of specific DNA barcodes and understanding of evolutionary history and conservation strategy in Family Syngnathidae.
Figure 1. ML phylogenetic tree of Halicampus grayi and other representative Syngnathidae species based on the complete mitochondrial protein coding sequences. Numbers above the lines indicate the bootstrap value for the ML analysis based on 100 replicates. The GenBank accession numbers were listed following the species name.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen M, Zhu L, Chen J, Zhang G, Cheng R, Ge Y. 2018. The complete mitochondrial genome of the short snouted seahorse Hippocampus hippocampus, Linnaeus 1758 (Syngnathiformes: Syngnathidae) and its phylogenetic implications. Conserv Genet Resour. 10:783–787.
- Dawson CE. 1982. Synopsis of the Indo-Pacific genus Solegnathus (Pisces: Syngnathidae). Jpn J Ichthyol. 29:139–161.
- Fang Y, Zhu L, Chen M, Ge Y, Zhang G, Cheng R. 2018. Characterization of the complete mitochondrial genome of the medical pipefish doryichthys boaja bleeker 1850. Mitochondrial DNA B Resour. 3:881–883.
- Gao L, Yin Y, Li J, Yuan Y, Jiang C, Gao W, Huang L. 2018. Identification of traditional Chinese medicinal pipefish and exclusion of common adulterants by multiplex PCR based on 12S sequences of specific alleles. Mitochondrial DNA A: DNA Mapp Seq Anal. 29:340–346.
- Garcia E, Rice CA, Eernisse DJ, Forsgren KL, Quimbayo JP, Rouse GW. 2019. Systematic relationships of sympatric pipefishes (Syngnathus spp.): a mismatch between morphological and molecular variation. J Fish Biol. 95:999.
- Ge Y, Zhu L, Chen M, Zhang G, Huang Z, Cheng R. 2018. Complete mitochondrial genome sequence for the endangered Knysna seahorse Hippocampus capensis Boulenger 1900. Conservation Genet Resour. 10:461–465.
- Lai M, Sun S, Chen J, Lou Z, Qiu F, Zhang G, Cheng R. 2019. Complete mitochondrial genome sequence for the seahorse adulteration Hippocampus camelopardalis Bianconi 1854. Mitochondrial DNA B Resour. 4:432–433.
- Sun S, Lai M, Huang Z, Ge Y, Zhang G, Cheng R. 2019. Characterization of the complete mitochondrial genome sequence of medical spiny pipehorse Solegnathus spinosissimus and its phylogenetic analysis. Mitochondrial DNA B Resour. 4:1772–1773.
- Vincent ACJ, Foster SJ, Koldewey HJ. 2011. Conservation and management of seahorses and other Syngnathidae. J Fish Biol. 78:1681–1724.
- Wang X, Zhang Y, Zhang H, Qin G, Lin Q. 2019. Complete mitochondrial genomes of eight seahorses and pipefishes (Syngnathiformes: Syngnathidae): insight into the adaptive radiation of syngnathid fishes. BMC Evol Biol. 19:119.
- Zhang H, Zhang Y, Qin G, Lin Q. 2015. The complete mitochondrial genome sequence of the network pipefish (Corythoichthys flavofasciatus) and the analyses of phylogenetic relationships within the Syngnathidae species. Mar Genomics. 19:59–64.
- Zhu L, Chen M, Cheng R, Ge W, Zhang G, Ge Y. 2019. Complete mitochondrial genome characterization of the alligator pipefish Syngnathoides biaculeatus and phylogenetic analysis of family Syngnathidae. Conserv Genet Resour. 11:401–404.
