Abstract
Bombyx mandarina is generally thought to be the wild ancestor nearest to the domesticated silkworm, Bombyx mori. Here, we report the complete mitochondrial genome (mitogenome) of B. mandarina strain Shiquan. The mitogenome contains 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one A + T-rich region. Phylogenetic analysis provides solid evidence that B. mandarina Shiquan belongs to Chinese B. mandarina, rather than Japanese B. mandarina. The new mitogenome provides useful information to further explore the origin and domestication of this species.
The wild silkworm, Bombyx mandarina, is believed to be the ancestor nearest to the domesticated silkworm Bombyx mori (Goldsmith et al. Citation2005; Xia et al. Citation2014). Extant in Asian mulberry fields, B. mandarina has two main types, which differ in chromosome number. Bombyx mandarina living in Japan has 27 chromosomes per haploid genome, while B. mandarina in China carries 28 chromosomes in its haploid genome, equal to those of B. mori (Banno et al. Citation2004; Nakamura et al. Citation1999). In the present study, we report the complete mitogenome sequence of B. mandarina strain Shiquan, thus providing a solid evidence that B. mandarina Shiquan has a close relationship with the B. mandarina from northern of China.
Bombyx mandarina was collected from Shiquan, Shaanxi province, China (N32°56′3.52″, E108°48′2.35″). After morphological identification, the specimens were successively sub-cultured by Shaanxi Key Laboratory of Sericulture, Ankang University (N32°41′55.93″, E108°58′45.83″), Ankang, China. Total genomic DNA was extracted from a single pupa using a standard phenol-chloroform method. The specimen and its DNA are stored with the archival number of AKWS_30 in Shaanxi Key Laboratory of Sericulture, Ankang University. A 400 bp insertion DNA library was constructed and performed paired-end using the Illumina Miseq platform (Illumina Inc., San Diego, CA). A5-miseq V20150522 (Coil et al. Citation2015) and SPAdesv3.9.0 software (Bankevich et al. Citation2012) were used to assemble the obtained high-quality paired-end reads. Genome annotation was performed using MITOS (Bernt et al. Citation2013) and tRNAscan-SE server (Schattner et al. Citation2005).
The complete mitogenome of B. mandarina is 15,662 bp in length (GenBank accession no. MN400656) and contains 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and 1 A-T-rich region. The overall base composition was estimated to be 43.16%A, 38.30%T, 11.28%C, and 7.30%G, with a high A-T content of 81.46%, which is similar to that of B. mori (Zhang et al. Citation2016). All PCGs begin with ATN, except for COX1 gene starting with CGA. Eleven PCGs use TAA as the stop codon, whereas COX1 and COX2 end with a single T. All of the 22 tRNA genes, ranging in size from 64 to 75 bp, have a typical cloverleaf secondary structure. The gene order and orientation of B. mandarina were consistent with those of B. mori (Li et al. Citation2016; Zhang et al. Citation2016).
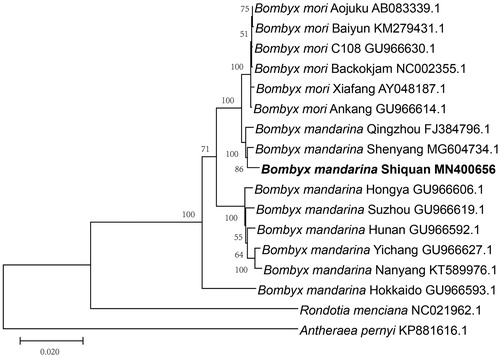
The phylogenetic relationship was recovered using the Maximum-Likelihood method in MEGA 7.0 (Kumar et al. Citation2016), based on complete mitochondrial genomes including strains of B. mandarina and B. mori. Phylogenetic analysis revealed that B. mandarina Shiquan belongs to Chinese B. mandarina and has close relationship with the wild silkworm from northern of China (). The mitogenome presented here will provide useful information for evolutionary studies of the origin of the present-day silkworm.
Disclosure statement
The authors report no conflict of interest and are responsible for the content and writing of the article.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Banno Y, Nakamura T, Nagashima E, Fujii H, Doira H. 2004. M chromosome of the wild silkworm, Bombyx mandarina (n = 27), corresponds to two chromosomes in the domesticated silkworm, Bombyx mori (n = 28). Genome. 47:96–101.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31:587–589.
- Goldsmith MR, Shimada T, Abe H. 2005. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol. 50:71–100.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li F, Zhang H, Liu P, Wang Y, Meng Z. 2016. Complete mitochondrial genome of a hybrid strain of the domesticated silkworm (Qiufeng x Baiyu). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1844–1845.
- Nakamura T, Banno Y, Nakada T, Nho SK, Xu MK, Ueda K, Kawarabata T, Kawaguchi Y, Koga K. 1999. Geographic dimorphism of the wild silkworm, Bombyx mandarina, in the chromosome number and the occurrence of a retroposon-like insertion in the arylphorin gene. Génome. 42:1117–1120.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Xia Q, Li S, Feng Q. 2014. Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu Rev Entomol. 59:513–536.
- Zhang H, Li F, Zhu X, Meng Z. 2016. The complete mitochondrial genome of Bombyx mori strain Baiyun (Lepidoptera: Bombycidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1652–1653.