Abstract
Flavoperla is a small genus in Acroneuriinae, which was established by Chu but was treated as a synonym for a long time until Uchida and Harper recognized Flavoperla as a valid genus. This genus includes 15 species in the world now, including six species from China. Here, we sequenced and annotated the mitochondrial genome of Flavoperla sp. under the next-generation sequence technology, the first representative of Genus Flavoperla. The complete mitochondrial genome of Flavoperla sp. is 15,796 bp in length with an A + T content of 68.3% showed a positive AT-skew (0.037) and a negative GC-skew (−0.262). The gene order and organization of the mitochondrial genome is consistent with other stoneflies. The control region had the highest A + T content. Eleven PCGs started with the typical codon (ATN), the remaining PCGs started with TTG (ND1and ND5). All PCGs terminated with TAA/TAG, except COII and ND5 used single T. The phylogenetic tree by maximum likelihood (ML) and Bayesian (BI) methods supported that Flavoperla sp. was the sister group to Niponiella limbatella.
The subfamily Acroneuriinae of Perlidae contains more than 500 species worldwide (DeWalt et al. Citation2019). Flavoperla is a small genus in Acroneuriinae that was initially established by Chu (Citation1929) but was treated as a synonym of genus Gibosia by Wu (Citation1935) for a long time until Uchida (Citation1990) and Harper (Citation1994) recognized Flavoperla as a valid genus. Recently, three species were described and reported from Southwestern and Northwestern China. This genus includes 15 species in the world, including six species from China (Chen Citation2019; DeWalt et al. Citation2019; Liu et al. Citation2019). Only five mitochondrial genomes in the Acroneuriinae were sequenced and published to date (Huang et al. Citation2015; Cao, Li et al. Citation2019; Cao, Wang, Li et al. Citation2019; Cao, Wang, Zhang et al. Citation2019; Li et al. Citation2019). Here, we sequenced and annotated the mitochondrial genome of Flavoperla sp. under the next-generation sequence technology, the first representative of Genus Flavoperla and reconstructed the phylogenetic position of Flavoperla. The specimen of Flavoperla sp. was collected in Huangshan Mountain, Anhui Province, China in 2016 (30.077°N, 118.164°E) by Cao Yuyan. The samples and voucher specimens (No. Voh-0120) were stored in 100% ethanol in the field and then stored at −20 °C in Henan institute of Science and Technology (HIST), China. Genomic DNA was extracted from the adult muscle tissues by the Qiagen DNAeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) stored at −20 °C until it was sent to sequencing. The base composition was analyzed by MEGA version 7.0.14 (Kumar et al. Citation2016). We calculated the base composition by using the formulas AT and GC skew (AT skew=(A − T)/(A + T), GC skew=(G − C)/(G + C)) (Perna and Kocher Citation1995).
The complete mitochondrial genome of Flavoperla sp. is a double-stranded closed-circular molecule of 15,796 bp in length with the GenBank accession number MN419916. Similar to other published stoneflies, the gene order, and organization of the mitochondrial genome (contains 37 genes and a control region)is consistent with those of putative ancestor of insects (Boore Citation1999). The mitochondrial genome of Flavoperla sp. with an A + T content of 68.3% showed a positive AT-skew (0.037) and a negative GC-skew (–0.262). The control region possessed the highest A + T content (72.7), and the A + T content of the 13 PCGs, 22 tRNAs, 2 rRNAs was 67.1, 70.6, and 70.9%, respectively. Eleven PCGs started with the typical codon (ATN), the remaining two PCGs started with TTG (ND1and ND5). All PCGs terminated with TAA or TAG, except COII and ND5 used the incomplete T as stop codon.
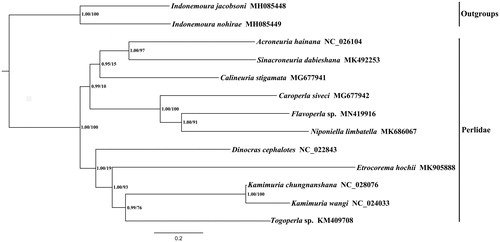
To better understand the phylogenetic relationship of the Flavoperla sp. with other closely related species, we conducted the sequences of PCG and two rRNAs among from eleven Perlidae and two Nemouridae species (as outgroups) by maximum likelihood (ML) and Bayesian (BI) methods to obtain the phylogenetic tree. The phylogenetic tree () in this study was consistent with the proposed and widely accepted view of the internal relationships of the Flavoperla sp. was sister clade to Niponiella limbatella.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Cao JJ, Li WH, Wang Y. 2019. The complete mitochondrial genome of a Chinese endemic stonefly species, Sinacroneuria dabieshana (Plecoptera: Perlidae). Mitochondr DNA B Resour. 4(1):1327–1328.
- Cao JJ, Wang Y, Li N, Li WH, Chen X. 2019. Characterization of the nearly complete mitochondrial genome of a Chinese endemic stonefly species, Caroperla siveci (Plecoptera: Perlidae). Mitochondr DNA B Resour. 4(1):553–554.
- Cao JJ, Wang Y, Zhang GQ, Yi SQ, Li WH. 2019. The characterization of the mitochondrial genome of Calineuria stigmatica (Plecoptera: Perlidae). Mitochondr DNA B Resour. 4(2):2796–2797.
- Chen ZT. 2019. On the identity of the genus Flavoperla (Plecoptera: Perlidae), with description of a new species in southwestern China. Zootaxa. 4613(1):127–134.
- Chu YT. 1929. Descriptions of four new species and one new genus of stone-flies in the family Perlidae from Hangchow. China J. 10:88–92.
- DeWalt RE, Maehr MD, Neu-Becker U, Stueber G. 2019. Plecoptera species file online. [accessed 2019 Sep 19]. http://Plecoptera.SpeciesFile.org.
- Harper PP. 1994. Plecoptera. In: Morse JC, Yang L, Tian L, editors. Aquatic insects of China useful for monitoring water quality. Nanjing, China: Hohai University Press; p. 176–209.
- Huang MC, Wang YY, Liu XY, Li WH, Kang ZH, Wang K, Li XK, Yang D. 2015. The complete mitochondrial genome and its remarkable secondary structure for a stonefly Acroneuria hainana Wu (Insecta: Plecoptera: Perlidae). Gene. 557(1):52–60.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li JF, Cao JJ, Wang Y, Kong FB. 2019. The mitochondrial genome analysis of a stonefly, Niponiella limbatella (Plecoptera: Perlidae). Mitochondr DNA B Resour. 4(1):1666–1667.
- Liu H, Yan Y, Li W. 2019. Two new species of Flavoperla (Plecoptera: Perlidae) from Shaanxi Province of China. Zootaxa. 4614(2):395–400.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at four-fold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358.
- Uchida S. 1990. A revision of the Japanese Perlidae (Insecta: Plecoptera), with special reference to their phylogeny [unpublished PhD thesis]. Tokyo, Japan: Tokyo Metropolitan University; p. 228.
- Wu CF. 1935. Order IX. Plecoptera. Vol. 1. In: Wu CF, editor. Catalogus insectorum sinensium. Peiping, China: Fan Memorial Institute of Biology; p. 299–315.