Abstract
The complete mitogenome of Sphaeroma sp. (Crustacea, Isopod, Sphaeromatidae) was determined in this study. The total length of mitogenome was 15,839 bp, and contained 13 protein-coding genes, 21 tRNA genes, 2 rRNA genes, 1 control region, and 2 unknown fragments which longer than 200 bp. The nucleotide composition was 25.80% A, 16.56% C, 25.94% G and 31.67% T. Six initiation codons (ATA, ATC, ATG, ATT, ACG, GTG) and three termination codons (TAA, TAG, TA–) were used in the protein-coding genes. The length of tRNA genes ranged from 52 to 65 bp, and tRNA-Arg was not identified. The phylogenetic result showed Sphaeroma sp. was closely related to Sphaeroma terebrans with high bootstrap value supported.
Sphaeromatidae is one of the largest families in Isopod (979 species in 125 genera) (WoRMS Editorial Board Citation2019). A part of Sphaeromatidae genetic data had been sequenced, nonetheless, only one complete mitochodrial genome was published in this family, which was Sphaeroma terebrans collected from China (Yang et al. Citation2019). Here, we determined the complete mitogenome of Sphaeroma sp. and analyzed the phylogenetic position of this species, hoping that could contribute to the molecular and phylogenetic study about isopod crustaceans.
Specimens of Sphaeroma sp. were collected from Beihai Coastal National Wetland Park, Beihai, Guangxi Province, China (21°24′41.11″N, 109°10′23.26″E) on October 2016 and preserved in 95% ethanol. After morphological identification, the specimen was deposited in Guangxi Mangrove Research Center with a voucher number DG20161009. The experimental protocol, data analysis methods and Maximum likelihood (ML) phylogenetic reconstruction followed Yang et al. (Citation2019). The outgroup Metacrangonyx longipes and 12 species of Isopod mitogenomes available in GenBank were used for the phylogenetic reconstruction.
The complete mitogenome of sphaeroma sp. was 15, 839 bp in length (GenBank accession number MN263919) and its overall base composition was 25.80% A, 16.56% C, 25.94% G, and 31.67% T. The mitogenomic variation between Sphaeroma sp. and S. terebrans (MK460228) was 28.3%. It consisted of 13 protein-coding genes, 21 transfer RNA genes, two rRNA genes, one control region and two unknown fragments which longer than 200 bp. Despite extensive efforts to find secondary structures in control region and unknown fragments, the tRNA-Arg gene was not identified in the mitogenome. Except for Cytb, ND1, ND4, and ND4L were encoded on the L-strand, the remaining 9 protein-coding genes were encoded on the H-strand. This arrangement of protein-coding genes was similar to Ligia oceanica (Kilpert and Podsiadlowski Citation2006) and S. terebrans (Yang et al. Citation2019). It was important to note that three protein-coding genes started with ATG codon (ND4, ATP6, and COX3), four with ATT codon (ND4L, ND6, ND2, and ATP8), two with ATA codon (ND1 and ND5), two with ATC codon (COX2 and Cytb), one with GTG codon (ND3), and one with ACG codon (COX1). All the protein-coding genes used TAA, TAG, and TA– as termination codons. The 21 tRNA genes interspersed between the rRNAs and protein-coding genes, ranging from 52 to 65 bp. The control region was 813 bp, presenting a high A + T content (62.12%).
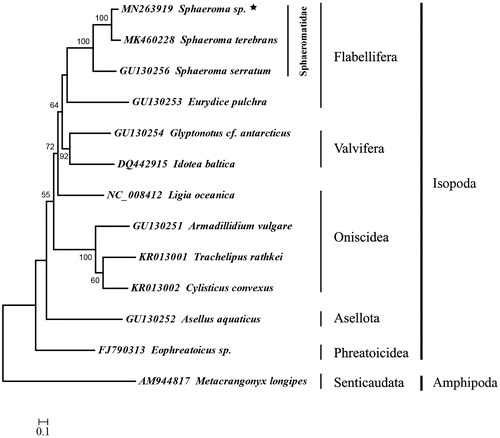
Five suborders of the Isopod were showed in the ML tree and most nodes were well supported (). The suborder Phreatoicidea was the basal clade. The Flabellifera was sister to the Valvifera. Within the family Sphaeromatidae of the suborder Flabellifera, Sphaeroma sp. clustered to S. terebrans, and then S. serratum clustered to this clade. Ligia oceanica clustered to the clades of Valvifera and Flabellifera instead of Oniscidea, it suggested that the suborder Oniscidea was a polyphyletic group. In conclusion, the complete mitogenome of Sphaeroma sp. could provide essential and important DNA molecular data for further studies of Isopod phylogeny.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kilpert F, Podsiadlowski L. 2006. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genom. 7(1):241.
- WoRMS Editorial Board. 2019. Word Register of Marine Species [Internet]; [accessed 2019 Sep 25]. Available from http://www.marinespecies.orgatVLIZ.
- Yang M, Gao T, Yan B, Chen X, Liu W. 2019. Complete mitochondrial genome and the phylogenetic position of a wood-boring Isopod Sphaeroma terebrans (Crustacea, Isopod, Sphaeromatidae). Mitochondrial DNA Part B. 4(1):1920–1921.