Abstract
The generitype Lessonia flavicans Bory is an endemic and important kelp from Sub-Antarctic Magellanic ecoregion that shows affinity to extreme salinity, temperature, and photoperiod conditions. Genomic analysis of L. flavicans from Rinconada Bulnes, Punta Arenas, Chile, resulted in the assembly of its organellar genomes. The L. flavicans complete mitogenome is 37,226 base pairs (bp) in length and contains 66 genes (GenBank accession number MN561186), the complete plastid genome is 130,085 bp and has 173 genes (MN561187) and the data assembled 8205 bp of the nuclear ribosomal cistron (MN561188). The organellar genomes are similar in structure and content to L. spicata (Suhr) Santelices and other Laminariales.
Lessonia flavicans is an intertidal to shallow subtidal kelp distributed in the Beagle Channel (Chile), Puerto Deseado (Argentina), and the Falkland Islands (Searles Citation1978; Mansilla et al. Citation2014). This species is characterized as having a stipe somewhat treelike cylindrical, branches compressed, blades oval-linear, somewhat denticulated, and yellowish (Searles Citation1978; Asensi and de Reviers Citation2009). Lessonia flavicans have major ecological roles in the structuring of benthic marine communities and are commercially exploited for the extraction of the phycocolloid alginate (Mansilla et al. Citation2014). This study characterized the complete organellar genomes of L. flavicans from Rinconada Bulnes, Punta Arenas, Chile to determine its genomic structure and genetic relationship as the generitype of the genus Lessonia.
DNA was extracted from L. flavicans (Specimen Voucher- LEMAS0005) using the NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions. The 150 bp PE Illumina library construction and sequencing were performed by myGenomics, LLC (Alpharetta, GA). The genomes were assembled using default de novo settings in CLC Genomics Workbench version 12.0 (QIAGEN Bioinformatics, Redwood City, CA) and Geneious Prime to close gaps (Biomatters, Ltd, Auckland, New Zealand). The genes were annotated manually using blastx, NCBI ORFfinder, and tRNAscan-SE version 1.21 (Schattner et al. Citation2005). The L. flavicans mitogenome was aligned to other mitogenomes using MAFFT (Katoh and Standley Citation2013). The phylogenetic analysis was executed with RAxML-NG (Kozlov et al. Citation2019) using the GTR + gamma model and 1,000 bootstraps. The tree was visualized with TreeDyn version 198.3 at Phylogeny.fr (Dereeper et al. Citation2008).
The mitogenome of L. flavicans is 37,226 bp in length and contains 66 genes. It is A + T rich (66.3%) and includes 25 tRNA (trnK and trnS occur in duplicate, trnL and trnM in triplicate), 17 ribosomal proteins, 3 rRNA (rnl, rns, and rrn5), 3 orfs (orf41, orf129, and orf378), and 18 other genes involved in electron transport and oxidative phosphorylation. The plastid genome of L. flavicans is 130,085 bp and contains 173 genes. It is A + T biased (69.2%) and includes 45 ribosomal proteins, 27 tRNA (trnA, trnG, trnI, trnR, and trnS occur in duplicate, trnM occurs in triplicate), 27 photosystem I and II, 20 ycf, 8 cytochrome b/f complex, 8 ATP synthase, 4 RNA polymerase, 6 rRNA, and 28 other genes. The mitogenome and plastid genome of L. flavicans are similar in length, content, and organization to other Laminariales (Chen et al. Citation2019; Tineo et al. Citation2019; Zheng et al. Citation2019).
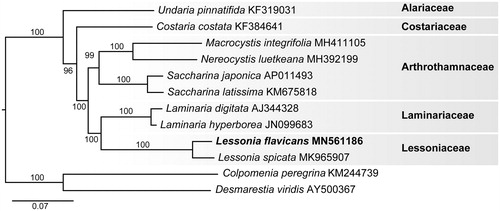
Phylogenetic analysis of the L. flavicans mitogenome resolved it in a fully supported clade with L. spicata (). The mitogenome of L. flavicans differed in pairwise distance from L. spicata by 3.8%. The plastid genome of L. flavicans differed from L. spicata by 1.4%. The evolutionary relationship of the generitype L. flavicans confirms that Lessoniaceae is closely allied with Laminariaceae (Tineo et al. Citation2019).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Asensi A, de Reviers B. 2009. Illustrated catalogue of types of species historically assigned to Lessonia (Laminariales, Phaeophyceae) preserved at PC, including a taxonomic study of three South-American species with a description of L. searlesiana sp. nov. and a new lectotypification of L. flavicans. Cryptogamie Algol. 30:9–249.
- Chen J, Zang Y, Shang S, Tang X. 2019. The complete mitochondrial genome of the brown alga Macrocystis integrifolia (Laminariales, Phaeophyceae). Mitochondrial DNA B. 4(1):635–636.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21): 4453–4455.
- Mansilla A, Rosenfeld S, Rendoll J, Murcia S, Werlinger C, Yokoya N, Terrados J. 2014. Tolerance response of Lessonia flavicans from the sub-Antarctic ecoregion of Magallanes under controlled environmental conditions. J Appl Phycol. 26(5):1971–1977.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 33:686–689.
- Searles RB. 1978. The genus Lessonia Bory (Phaeophyta, Laminariales) in southern Chile and Argentina. Br Phycol J. 13(4):361–381.
- Tineo D, Rubio KB, Melendez JB, Mendoza JE, Silva JO, Perez J, Esquerre EE, Perez-Alania M, Fernandez SL, Aguilar SE, et al. 2019. Analysis of the complete organellar genomes of the economically valuable kelp Lessonia spicata (Lessoniaceae, Phaeophyceae) from Chile. Mitochondrial DNA B. 4(2):2581–2582.
- Zheng Z, Chen H, Wang H, Jiang W, Cao Q, Du N. 2019. Characterization of the complete mitochondrial genome of bull kelp. Mitochondrial DNA B. 4(1):630–631.