Abstract
The complete mitogenome of Smaragdina nigrifrons (GenBank accession number MN584924) is 15,903 bp in length, and includes 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes, and a control region. The overall base composition was as follows: A, 38.18%; T, 35.25%; C, 15.97%; and G, 10.60%, with a total of A + T content of 73.43%. Eleven reading frame overlaps and thirteen intergenic regions were found in the mitogenome of S. nigrifrons. All 13 PCGs are initiated with the typical ATN codons, and are terminated with either the complete TAA/TAG codons or a single T residue. All tRNAs possess the typical cloverleaf secondary structures except for trnS1 (AGN). Phylogenetic analyses showed that S. nigrifrons was closely related to Cucujus clavipes, which was consistent with the conventional taxonomy.
The leaf beetle, Smaragdina nigrifrons (Coleoptera: Eumolpidae), is a major agricultural pest and distributed widely in China. The beetle cause severe economic damage to many plants, including jujube, corn, bean, abacus, and millet. The adults of S. nigrifrons feed on plant leaves and bite them into holes and notches (Gao et al. Citation2019). In this study, the specimen of S. nigrifrons were collected from Fanjing Mountain in Guizhou Province of China (N27°53′, E108°48′), and stored in 95% ethanol and registered in the Insect Specimen Room of Guiyang University with an accession number GYU-Col-2019001.
The complete mitogenome sequence of S. nigrifrons (GenBank accession number MN584924) is 15,903 bp in length, harboring the typical set of 37 mitochondrial genes which include 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (16S rRNA and 12S rRNA), and a control region (Boore Citation1999). All genes have the similar strands and location with that of click beetle, Agriotes hirayamai (Lin et al. Citation2018). The overall base composition of S. nigrifrons mitogenome was as follows: A, 38.18%; T, 35.25%; C, 15.97%; and G, 10.60%, with a total of A + T content of 73.43%. The AT-skew and GC-skew of this genome were 0.040 and −0.202, respectively. Amongst the 37genes, 14 genes (trnQ, trnC, trnY, trnF, trnH, trnP, trnL1, trnV, nad1, nad4, nad4L, nad5, 16S rRNA, and 12S rRNA) were located on the light strand (L-strand), while the remaining 23 genes were encoded on the heavy strand (H-strand).
The total length of 13 PCGs of S. nigrifrons was 11,034 bp. The 13 PCGs in the mitogenome initiated with the typical ATN as start codons (ATG for cox2, cox3, atp6, atp8, nad4, nad5, and cob; ATT for nad3, nad4L, nad6, and cox1; ATA for nad1 and nad2). Ten PCGs used typical termination codons TAA and TAG in S. nigrifrons, while only three PCGs (cox3, nad4, and nad5) stop with the incomplete termination signal T. There were 11 overlap regions comprising a total length of 212 bp and the largest spacer (92 bp) resided between trnA and trnN, and 13 intergenic spacer regions ranging in length from 1 to 8 bp, comprising a total length of 42 bp. The length of the tRNAs varied between 62 bp (trnE and trnT) and 70 bp (trnK), comprising a total length of 1447 bp. All 22 tRNAs possessed the typical cloverleaf structure except for trnS1 (AGN), which lacked the dihydrouridine arm and occurred commonly in most insects (Ohtsuki et al. Citation2002; Yuan et al. Citation2016). The 16S rRNA and 12S rRNA were 1283 and 740 bp in length, with the A + T contents of 77.71 and 73.51%, respectively. The control region was 1229 bp in length and had a remarkably high A + T content (81.86%), which was located between 12S rRNA and trnI.
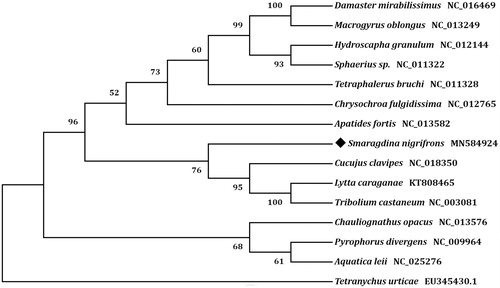
Phylogenetic analyses were performed with respect to the concatenated amino acid sequences of 13 PCGs. A neighbor-joining tree was constructed by the program MEGA version 7.0 (Kumar et al. Citation2016). The result showed that S. nigrifrons was closely related to Cucujus clavipes (), which was consistent with the conventional taxonomy.
Figure 1. Phylogenetic tree showing the relationship between Smaragdina nigrifrons and 13 other beetles based on neighbor-joining method. Beetle determined in this study was labeled with black diamond. Tetrancychus urticae was used as an outgroup. GenBank accession numbers of each species were listed in the tree.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication. The authors also are responsible for the content and writing of the article.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Gao SH, Lu CK, Jia YX, Zhao CM, Cao X. 2019. A new pest in vineyard under grass management model: study on the biological characteristics and control of Smaragdina nigrifrons. J Fruit Sci. 36:1185–1193.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger dataset. Mol Biol Evol. 33(7):1870–1874.
- Lin AL, Zhao XC, Song N, Zhao T. 2018. Analysis of the complete mitochondrial genome of click beetle Agriotes hirayamai (Coloeptera: Elateridae). Mitochondr DNA B. 3(1):290–291.
- Ohtsuki T, Kawai G, Watanabe K. 2002. The minimal tRNA: unique structure of Ascaris suum mitochondrial tRNA(Ser)(UCU) having a short T arm and lacking the entire D arm. FEBS Lett. 514(1):37–43.
- Yuan ML, Zhang QL, Zhang L, Guo ZL, Liu YJ, Shen YY, Shao RF. 2016. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol Phylogenet Evol. 104:99–111.
