Abstract
Habenaria ciliolaris is a kind of orchid with ornamental value. In this study, we reported the complete chloroplast genome of H. ciliolaris. The complete chloroplast genome is 154,544 bp in length, consists of a pair of inverted repeat (IR, 25,455 bp) regions, a large single-copy region (LSC, 84,032 bp) and a small single-copy region (SSC, 19,602 bp). It contains 179 genes, including 133 protein-coding genes, 38 tRNAs, and 8 rRNAs. A maximum-likelihood phylogenetic analysis demonstrated a close relationship between H. ciliolaris and Habenaria radiata. This work will be valuable for genetic and phylogenetic studies on H. ciliolaris.
Habenaria is one of the largest genera in the Orchidaceae family, including about 876 species and mainly distributed in tropical and subtropical areas (Govaerts et al. Citation2013). According to Flora of China, 58 species of Habenaria were reported in China (Zhang et al. Citation2016). The genus Habenaria is not only used as important medicinal plant in China but also has a high ornamental value (Tang Citation2016). Habenaria ciliolaris is a species belonging to the Habenaria genera. It grows on a hillside or under the forest at an altitude of 140–1800 m (Chen et al. Citation2009). However, few data are available regarding the H. ciliolaris chloroplast genome. Here, we reported the complete chloroplast genome sequence of H. ciliolaris. Our data will be valuable for genetic and phylogenetic studies on H. ciliolaris.
The plant material of H. ciliolaris was collected from Qinglong waterfall scenic area, Yongtai, Fujian province, China (25°46′23.30″N, 118°57′50.50″E). The voucher specimen is kept at the Herbarium of Fujian Agriculture and Forestry University (specimen code YT-MTYFH).
Total genomic DNA was extracted from fresh leaves using the cetyltrimethylammonium bromide (CTAB) protocol (Doyle and Doyle Citation1987) and chloroplast genome sequences were analyzed using Illumina Hiseq 2000; raw data were filtered with the NGS QC Toolkit (Patel and Jain Citation2012); the clean reads were compared with the chloroplast sequence of Habenaria daitata (GenBank accession no. NC.035834). We used Version 1.3 for SOAPdenovo assembly (Luo et al. Citation2012). After assembled, the obtained scaffolds and contigs were assembled into the chloroplast genome by Geneious version 2019.1.1 (Li et al. Citation2019), then Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004) was used to annotate chloroplast genome. The complete chloroplast genome of H. ciliolaris was submitted to GenBank with the accession number MN.495954.
The complete chloroplast genome of H. ciliolaris is 154,544 bp in size, which is composed of a large single-copy (LSC), a small single-copy (SSC), and two inverted repeat (IR) regions of 84,032, 19,602, and 25,455 bp, respectively. It contains 133 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The complete genome GC content was 37.9%.
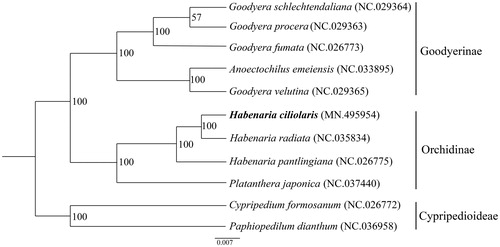
Phylogenetic analysis was conducted with 11 complete chloroplast genome sequences of Orchidaceae (Goodyera schlechtendaliana, Goodyera procera, Goodyera fumata, Anoectochilus emeiensis, Goodyera velutina, H. ciliolaris, Habenaria radiata, Habenaria pantlingiana, Platanthera japonica, Cypripedium formosanum, and Paphiopedilum dianthum), using a maximum-likelihood analysis of MEGA 6.0 (Tamura et al. Citation2013) with 1000 bootstrap replicates. The phylogenetic tree indicated that H. ciliolaris was most closely related to H. radiata, as expected ().
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Additional information
Funding
References
- Chen SC, Liu ZJ, Zhu GH, Lang KY, Tsi ZH, Luo YB, Jin XH, Cribb PJ, Wood JJ, Gale SW, et al. 2009. Flora of China. Vol. 25. Beijing (China): Science Press.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Govaerts R, Bemet P, Kratochvil K, Gerlach G, Carr G, Alrich P, Pridgeon AM, Pfahl J, Campacci MA, Baptista DH, et al. 2013. World checklist of Orchidaceae. London (UK): Botanic Gardens, Kew; [accessed 2013 Mar 14]. http://apps.kew.org/wcsp/.
- Li Y, Li Z, Hu Q, Zhai J, Liu Z, Wu S. 2019. Complete plastid genome of Apostasia shenzhenica (Orchidaceae). Mitochondr DNA B. 4(1):1388–1389.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1(1):18.
- Patel RK, Jain M. 2012. Ngs qc toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Tang H. 2016. DNA barcoding identification and ecological suitability of important medicinal plants of Orchidaceae. Ya'an (China): Sichuan Agricultural University; p. 9.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang WL, Gao JY, Liu Q. 2016. Habenaria chlorina, a newly recorded species of Orchidaceae form China. Plant Sci J. 34(1):18–20.