Abstract
In this study, the entire mitochondrial genome of Paragnetina indentata was sequenced. The circular mitochondrial genome was the first mitochondrial genome representing the genus Paragnetina, which consists of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 22 transfer RNA genes, and 1 control region with the length of 15,885 bp. In the complete sequence, the A + T content for 64.1% (A for 33.6%, T for 30.5%, C for 23.3%, and G for 12.7%). Among the 13 PCGs, 11 start codons is ATN and the start codon of ND1, ND2, and ND5 genes is TTG, GTG, and GTG, respectively. In addition, 11 of the PCGs used conservative termination codon TAA or TAG, except for COII and ND5 which terminated by the single T. By using the Bayesian inference (BI) and maximum-likelihood (ML) methods, the phylogenetic relationship showed that P. indentata was closely related to Togoperla sp. and the species of Perlinae were clustered in a clade.
There are 20 genera (including the genus Miniperla) in subfamily Perlinae of family Perlidae over the world (DeWalt et al. Citation2019). Over the past few decades, DNA sequencing and analysis have advanced rapidly, and phylogenetic inferences have been made at different levels of classification (Gibson et al. Citation2005; Bernt et al. Citation2013; Cameron Citation2014; Liu et al. Citation2014). As far as we know, seven complete mitochondrial genomes of Perlinae have been sequenced and published, but genus Paragnetina was not involved (Qian et al. Citation2014; Elbrecht et al. Citation2015; Wang, Ding, et al. Citation2016; Wang, Wang, et al. Citation2016; Chen et al. Citation2019; Liu et al. Citation2019; Zhang et al. Citation2019). We sequenced the complete mitochondrial genomes of Paragnetina indentata to provide supplementary information for future phylogenetic studies of Perlidae in this paper.
This specimen was collected by Xinyu Luo in Shaoguan, Guangdong Province, China (24.731°N, 114.267°E) on 2 February 2016. The specimen (No. VHL-0067) is now preserved in Henan Institute of Science and Technology (HIST), Henan Province, China. We extracted the total genomic DNA from the chest muscle of the adult by QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany), and sequenced the sample by high-throughput sequencing method.
The mitochondrial genome of P. indentata with a length of 15,885 bp was circular, which was deposited in NCBI with the accession number MN627431. Like other stoneflies, the genome composed 13 typical protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs), 22 transfer RNAs (tRNAs), and 1 control region (CR). The nucleotide composition of P. indentata is similar to that of other insect and it is inclined to A/T nucleotide, in which A accounted for 33.6%, T 30.5%, C 23.3%, G 12.7%, and A + T 64.1%. The AT skew and GC skew of this genome are 0.047 and −0.295, respectively. In addition, the A + T contents in PCGs, tRNAs, rRNAs, and CR were 61.9%, 68.3%, 68.0%, and 75.0%, respectively. All tRNAs have a typical clover secondary structure except the tRNAser (AGN) lacks dihydropyridine. The initial codon of 11 PCGs was ATN, and TTG, GTG, and GTG were used as start codon for ND1, ND2, and ND5. In addition, 11 PCGs used typical stop codon TAA/TAG, COII and ND5 used single T as termination. The length of lrRNA and srRNA was 1376 bp and 830 bp, with the content of A + T was 69.6% and 65.4%.
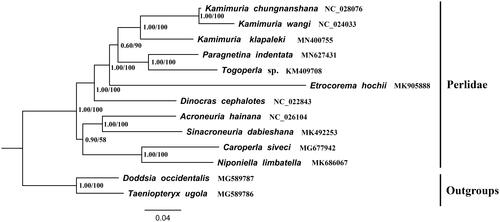
Based on the concatenated nucleotide sequences of PCG12 (including the first and second codon positions of PCGs) and two rRNAs from 11 Perlidae species and two species Doddsia occidentalis and Taeniopteryx ugola of Taeniopterygidae are used as outgroups for reestablished the phylogenetic relationship. Bayesian inference (BI) and maximum-likelihood (ML) were used for analysis, both two methods showed the same topology tree. As shown in , the phylogenetic tree indicated that P. indentata and Togoperla sp. were sister group, the clade P. indentata + Togoperla sp. was closely related to genus Kamimuria and the species of Perlinae were clustered in one clade. This result is consistent with traditional morphological classification (Sivec et al. Citation1988).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Bleidorn C, Braband A, Dambach J, Donath A, Fritzsch G, Golombek A, Hadrys H, Jühling F, Meusemann K, et al. 2013. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol. 69:352–364.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117.
- Chen JJ, Wang Y, Li WH, Cao JJ. 2019. Characterization of the complete mitochondrial genome of a stonefly species, Kamimuria klapaleki (Plecoptera: Pelidae). Mitochondrial DNA B Resour. 4(2):3416–3417.
- DeWalt RE, Maehr MD, Neu-Becker U, Stueber G. 2019. Plecoptera species file online; [accessed 2019 Oct 16]. http://Plecoptera.SpeciesFile.org.
- Elbrecht V, Poettker L, John U, Leese F. 2015. The complete mitochondrial genome of the stonefly Dinocras cephalotes (Plecoptera: Perlidae). Mitochondrial DNA A. 26(3):469–470.
- Gibson A, Gowri-Shankar V, Higgs PG, Rattray MA. 2005. A comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol Biol Evol. 22(2):251–264.
- Liu Y, Cox CJ, Wang W, Goffinet B. 2014. Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst Biol. 63(6):862–878.
- Liu ZZ, Wang Y, Li WH, Cao JJ. 2019. The complete mitochondrial genome of a stonefly species, Etrocorema hochii (Plecoptera: Pelidae). Mitochondrial DNA B Resour. 4(2):2690–2691.
- Qian YH, Wu HY, Ji XY, Yu WW, Du YZ. 2014. Mitochondrial genome of the stonefly Kamimuria wangi (Plecoptera: Perlidae) and phylogenetic position of Plecoptera based on mitogenomes. PLoS One. 9:e86328.
- Sivec I, Stark BP, Uchida S. 1988. Synopsis of the world genera of Perlinae (Plecoptera: Perlidae). Scopolia. 16:1–66.
- Wang K, Ding SM, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Kamimuria chungnanshana Wu, 1948 (Plecoptera: Perlidae). Mitochondrial DNA A. 27(5):3810–3811.
- Wang K, Wang YY, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Togoperla sp. (Plecoptera: Perlidae). Mitochondrial DNA A. 27:1703–1704.
- Zhang GQ, Wang Y, Cao JJ. 2019. The complete mitochondrial genome of a stonefly species, Neoperlops gressitti (Plecoptera: Pelidae). Mitochondrial DNA B Resour. 4(2):3324–3325.