Abstract
Platanthera minor is widely distributed in East Asia. The complete circular chloroplast genome with a length of 154,430 bp possesses the typical structure, consisting of a large single copy (LSC) of 83,536 bp, a small single copy (SSC) of 17,612 bp, and two inverted repeats (IR) of 26,641 bp. The average GC content of the genome is 36.7%. The circular P. minor chloroplast genome contains 114 genes, including 80 protein-coding genes, four rRNA genes, and 30 tRNA genes. The chloroplast sequence provided a resource for analyzing genetic diversity of the Orchidaceae family.
The Orchidaceae is one of the largest plant families, which is believed to contain more than 20,000 species (Dixon et al. Citation2003; Chase et al. Citation2015). Platanthera minor is a member of the genus Platanthera belonging to the subfamily Orchidoideae, widely distributed in East Asia. Platanthera minor mainly grows under forest on slopes, alpine meadows, with an altitude of 200 to 3000 m. So far, only two complete chloroplast genomes from the genus Platanthera has been reported (Dong et al. Citation2018; Lallemand et al. Citation2019). There have been no studies on the genome of P. minor. In this study, we reported the complete circular chloroplast genome sequence of P. minor and used the sequence for phylogenetic analysis.
Platanthera minor used for sequencing were collected from Tengchong Yunnan, China (25°0′06.9″N, 98°19′31.5″E). The voucher specimens (JXH10398) are deposited in the Herbarium, Institute of Botany, Chinese Academy of Sciences. The dry leaves were used to extract total DNA by CTAB (Li et al. Citation2013), and the complete chloroplast sequence was obtained using Illumina Hiseq 2500 and assembled after trimming using Geneious (Kearse et al. Citation2012). The DNA was deposited in State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China. The average coverage was 484.2. This sequence was annotated using PGA (Qu et al. Citation2019), and we manually checked the correction.
The circular P. minor chloroplast genome (GenBank accession MN416689) contains 114 genes, including 80 protein-coding genes, four rRNA genes, and 30 tRNA genes. Thirteen genes contain single intron but clpP, rps12 and ycf3 genes have two introns. The length of the complete chloroplast of P. minor is 154,430 bp, composed of a large single copy (LSC) of 83,536 bp, a small single copy (SSC) of 17,612 bp, and two inverted repeats (IR) of 26,641 bp. The average GC content of chloroplast is 36.7%.
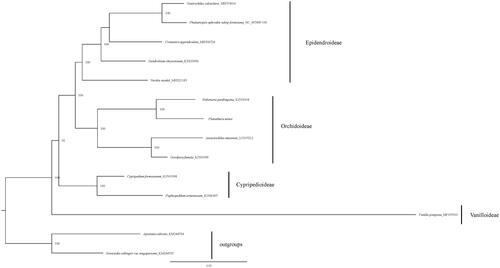
The phylogenetic tree was constructed based on the whole chloroplast genome of 13 species representing five orchid subfamilies. All protein-coding genes exported from Geneious (Kearse et al. Citation2012) and were aligned together with other representatives of Orchidoideae. We manually checked in BioEdit (Hall Citation1999) and concatenated these aligned sequences using SequenceMatrix (Vaidya et al. Citation2011) later. The phylogenetic analysis was performed using maximum likelihood method by IQtree with 1000 ultrafast bootstrap (Nguyen et al. Citation2015). The phylogenetic tree indicated that the P. minor is closer to Habenaria pantlingiana in Orchidoideae (). Our chloroplast sequence provided a resource for analyzing the genetic diversity of the Orchidaceae family.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Berg CVD, Schuiteman A. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177(2):151–174.
- Dixon KW, Kell SP, Barrett RL, Cribb PJ. 2003. Orchid conservation: a global perspective. Kota Kinabalu (Sabah): Natural History Publications.
- Dong W-L, Wang R-N, Zhang N-Y, Fan W-B, Fang M-F, Li Z-H. 2018. Molecular evolution of chloroplast genomes of orchid species: insights into phylogenetic relationship and adaptive evolution. IJMS. 19(3):716.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lallemand F, May M, Ihnatowicz A, Jąkalski M. 2019. The complete chloroplast genome sequence of Platanthera chlorantha (Orchidaceae). Mitochond DNA B Res. 4(2):2683–2684.
- Li JL, Wang S, Jing Y, Wang L, Zhou SL. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48:72–78.
- Nguyen L, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Qu X, Moore MJ, Li D, Yi T. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Vaidya GD, Lohman J, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.