Abstract
In the present study, the complete mitochondrial DNA sequence of the milkfish Lactarius lactarius is determined. The full-length of the mitochondrial genome consists of a 16,552 bp fragment, with the base composition of A (28.24%), C (29.82%), G (15.96%), and T (25.98%) with a high A + T content (54.22%). The base compositions present clearly the A-T skew, which is most obvious in the control region and protein-coding genes. It includes 2 ribosomal RNA (rRNA) genes, 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and a major non-coding control region (D-loop region). Furthermore, the composition and order of these genes are identical to most of other vertebrates. All the protein initiation codons are ATG, except that COX1 and ATP6 begin with GTG. All the protein termination codons are TAA, except ND3 and ND6 end with TAG, and COXII, ND4, Cytb finish only with incomplete T. Furthermore, the phylogenetic analysis base on 13 concatenated PCGs amino acid datasets among the 14 species, suggesting that high value support for the following sister clade with Toxotes chatareus. Our findings provide useful information for phylogenetic and evolutionary research of Perciformes species.
Keywords:
The false trevally Lactarius lactarius, is unique fish from Lactarius, Lactariidae, Perciformes. It distributes generally in the Indian Ocean-western and coastal area of China (Leis Citation1999). This species of habitat type is marine neritic and the extent and quality of habitat is ceaselessly decline in area. The present population trend of L. lactarius is unknown (Kaymaram et al. Citation2015) and is only known that partial of fish distributes in the Persian Gulf from a trawl sample off Iran (Valinassab et al. Citation2006). In the present study, we characterize the complete mitochondrial genome DNA (mtDNA) sequence of L. lactarius for phylogenetic analysis.
The samples of L. lactarius are collected from Hailing island, Yangjiang city, Guangdong province, China (111°52′00″E, 21°40′00″N). The specimens now are deposited in the South China Sea Fisheries Research Institute (Guangzhou, China). The genomic DNA extracts from standard phenol/chloroform method (Sambrook and Russel Citation2000). The method for preparing and sequencing the DNA library is referred to Zhu, Liang, et al. (Citation2017). For mtDNA sequence assemble, annotation and analysis are referred to Zhu, Wu, et al. (Citation2017). All the specimens (Accession number: Ssfri-F00156) and the genomic DNA are deposited in the Tropical and Subtropical Marine Life Museum of South China Sea Fisheries Research Institute, Guangzhou, Guangdong, China.
The complete mtDNA of L. lactarius is performed to be 16,552 bp in length (Accession number: MN604078), with the following nucleotide composition: A (28.24%), T (25.98%), C (29.82%), G (15.96%), and with a high A + T content (54.22%). Moreover, A (32.31%), T (34.08%), C (19.29%), and G (14.32%) are in D-loop region. There are ten overlapping regions, ranging from 1 to 10 bp. And 11 intergenic regions exist in this genome, ranging from 1 to 40 bp. Furthermore, the gene content is equally to other Perciformes mtDNA (Yagishita et al. Citation2009; Zhang et al. Citation2016; Zhu, Wu, et al. Citation2017), comprising 13 protein coding genes (PCGs), 22 tRNAs, 2 rRNAs, and a control region.
Only one PCGs (ND6), and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) are encoded by L strand, while two rRNA genes (12S rRNA and 16S rRNA), twelve PCGs (ND1, ND2, COX1, COXII, ATP8, ATP6, COXIII, ND3, ND4L, ND5, and Cytb), fourteen tRNA genes (Phe, Val, Leu, Ile, Met, Trp, Asp, Lys, Gly, Arg, His, Ser, Leu, and Thr), and a non-coding region (D-loop region) are encoded by H strand. Eleven of thirteen PCGs start with the representative initiation codon ATG, only COX1 and ATP6 with GTG. Eight PCGs initial TAA and ND3, ND6 use TAG as stop codon, whereas COXII, ND4, and Cytb use incomplete stop codon T-. It is analogously with that in Toxotes chatareus, Elagatis bipinnulata, and Lutjanus carponotatus mtDNA (Yagishita et al. Citation2009; Ma et al. Citation2017; Kim et al. Citation2019).
There are 22 tRNAs genes in the mtDNA of L. lactarius, ranging from 67 bp (tRNACys) to 75 bp (tRNALys). Non-coding region is A + T enrichment region, which is related to transcription and replication (Clayton Citation1992). The non-coding region of L. lactarius is located between tRNAPro and tRNAPhe genes, is 845 bp long with high A + T content (66.39%). Furthermore, all the tRNAs can be folded into the representative cloverleaf secondary structures, except tRNASer(GCT) which lacks dihydrouridine (DHU) arm.
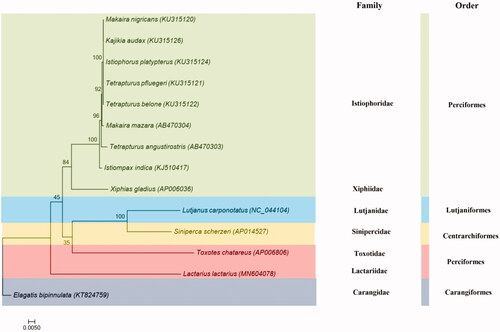
To investigate phylogenetic relationships between Centrarchiformes, Perciformes, Lutjaniformes, Carangiformes, and Istiophoriformes, a phylogenetic tree is constructed by MEGA 7.0 based on 13 tandem PCGs amino acid sequences with Maximum likelihood (ML) method and MtMam + I + G + F model (Kumar et al. Citation2016). Because of both two species are belong to Perciformes, T. chatareus is grouped with L. lactarius as the sister species (). The L. lactarius mtDNA will provide additional genetic information for genetic analyses in the future study.
Acknowledgements
We thank the editor and anonymous reviewers for providing valuable comments on the manuscript.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Clayton DA. 1992. Transcription and replication of animal mitochondrial DNAs. Int Rev Cytol. 141:217–232.
- Kaymaram F, Abdulqader E, Al-Husaini M, Hartmann S, Alam S, Almukhtar M, Alghawzi Q. 2015. Lactarius lactarius. The IUCN Red List of Threatened Species 2015: e.T46086218A57139758. Arizona: Arizona State University.
- Kim G, Lee JH, Alam MJ, Lee SR, Andriyono S. 2019. Complete mitochondrial genome of Spanish flag snapper, Lutjanus carponotatus (Perciformes: Lutjanidae). Mitochondrial DNA B Resour. 4(1):568–569.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Leis J. 1999. Lactariidae. In: Carpenter K. and Niem V. editors. The living marine resources of the western-central pacific. Rome: FAO; p. 2649.
- Ma C, Ma H, Zhang H, Feng C, Wei H, Wang W, Chen W, Zhang F, Ma L. 2017. The complete mitochondrial genome sequence and gene organization of the rainbow runner (Elagatis bipinnulata) (Perciformes: Carangidae). Mitochondrial DNA A DNA. 28(1):5–6.
- Sambrook JD, Russell W. 2000. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- Valinassab T, Daryanabard R, Dehghani R, Pierce GJ. 2006. Abundance of demersal fish resources in the Persian Gulf and Oman Sea. J Mar Biol Ass. 86(6):1455–1462.
- Yagishita N, Miya M, Yamanoue Y, Shirai SM, Nakayama K, Suzuki N, Satoh TP, Mabuchi K, Nishida M, Nakabo T. 2009. Mitogenomic evaluation of the unique facial nerve pattern as a phylogenetic marker within the percifom fishes (Teleostei: Percomorpha). Mol Phylogenet Evol. 53(1):258–266.
- Zhang DC, Wang L, Guo H, Ma Z, Zhang N, Lin J, Jiang SG. 2016. Complete mitochondrial genome of Florida pompano Trachinotus carolinus (Teleostei, Carangidae). Mitochondrial DNA A. 27(1):597–598.
- Zhu KC, Liang YY, Wu N, Guo HY, Zhang N, Jiang SG, Zhang DC. 2017. Sequencing and characterization of the complete mitochondrial genome of Japanese Swellshark (Cephalloscyllium umbratile). Sci Rep. 7(1):15299.
- Zhu KC, Wu N, Sun XX, Guo HY, Zhang N, Jiang SG, Zhang DC. 2017. Characterization of complete mitochondrial genome of fives tripe wrasse (Thalassoma quinquevittatum, Lay and Bennett, 1839) and phylogenetic analysis. Gene. 598:71–78.