Abstract
We report here the first mitogenome sequences for the chlorophyte class Chlorodendrophyceae. The mitogenomes of Tetraselmis sp. CCMP 881 and Scherffelia dubia (SAG 17.86) are 46,904 bp and 78,958 bp long, respectively, but their gene repertoires are almost identical. Each genome harbors an inverted repeat (IR). The 14,105-bp IR of S. dubia encodes seven genes in addition to a part of rps19, whereas the 2445-bp IR of Tetraselmis sp. CCMP 881 contains a single gene. Considering that an IR has also been found in the mitogenomes of certain earlier-diverging chlorophytes, the IRs of chlorodendrophycean algae probably represent ancestral features.
The Chlorodendrophyceae is a small class of green algae belonging to the Chlorophyta that comprises marine and freshwater scaly quadriflagellates (Massjuk Citation2006; Leliaert et al. Citation2012). In this study, we sequenced the mitogenomes of Tetraselmis sp. CCMP 881 and Scherffelia dubia (Chlorodendrales, Chlorodendraceae). These sequences are the first complete mitogenomes available for the Chlorodendrophyceae.
The S. dubia strain (SAG 17.86) was obtained from the Culture Collection of Algae at the University of Göttingen, whereas the Tetraselmis sp. strain (CCMP 881) was obtained from the Bigelow National Center for Marine Algae and Microbiota (Maine, USA). For DNA isolation, DNA sequencing and gene annotation, we used the methodologies we previously described for the plastomes of the same algal strains (Turmel et al. Citation2016). Briefly, random clone plasmid libraries were prepared from an A + T-rich DNA fraction and inserts of selected clones were sequenced using the Sanger method. Genomic regions, not represented in the clones analyzed, were sequenced from PCR fragments. All sequence data were generated by the “Plateforme d’Analyses Génomiques” of Laval University and they were edited and assembled using Sequencher 5.1 (Gene Codes Corporation, Ann Arbor, MI).
The Tetraselmis sp. and S. dubia mitogenomes were assembled as circular molecules of 46,904 bp (GenBank MN642087) and 78,958 bp (GenBank MN642088), respectively. Despite their important variation in size, their gene repertoires are almost identical, with 60 and 61 conserved genes, respectively (duplicated genes were counted only once). The Tetraselmis sp. mitogenome is lacking three tRNA genes found in S. dubia (trnC(gcc), R(acg), and T(ggu)), while the S. dubia mitogenome is lacking two ribosomal protein-coding genes found in Tetraselmis sp. (rps7 and rpl10). Gene order is highly rearranged between the two algal mitogenomes.
Each chlorodendrophycean genome harbors an inverted repeat (IR) but these IRs differ substantially in size and gene content. The 2445-bp IR of Tetraselmis sp. contains a single gene (trnMf(cau)), whereas the 14,105-bp IR of S. dubia contains eight genes (including all three rRNA genes) as well as most of the rps19 coding sequence. Considering that an IR has also been uncovered in the mitogenomes of five prasinophytes representing three distinct classes (Mamiellophyceae, Mamiellales (Robbens et al. Citation2007; Worden et al. Citation2009); Palmophyllophyceae, Prasinococcales (Pombert et al. Citation2013); Pyramimonadophyceae, Pyramimonadales (Satjarak et al. Citation2017)), it is likely that the IRs reported here represent ancestral features.
The Tetraselmis sp. and S. dubia mitogenomes comprise two and five group I introns, respectively. Both Tetraselmis sp. introns reside in rnl and encode a LAGLIDADG homing endonuclease, whereas the S. dubia introns lie within rns, rnl (two sites), and cox1 (two sites) and exhibit no ORF. One of the rnl introns is inserted at the same site in the two chlorodendrophycean algae.
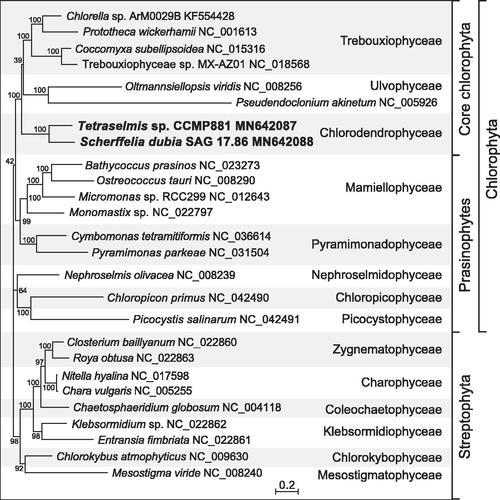
A maximum-likelihood phylogeny was inferred from 32 concatenated mitogenome-encoded proteins of 17 chlorophytes and 9 streptophyte green algae using RAxML v.8.2.3 (Stamatakis Citation2014). The best-scoring tree shows that the two representatives of the Chlorodendrophyceae form a strongly supported clade occupying a basal position within the core Chlorophyta ().
Figure 1. RAxML analysis of 32 concatenated mitogenome-encoded proteins from 17 chlorophytes and 9 streptophyte green algae. The figure shows the best-scoring tree, with the bootstrap support values (100 replicates) reported on the nodes. GenBank accession numbers are provided for the mitogenomes of all taxa. The scale bar denotes the estimated number of amino acid substitutions per site. The data set was generated using the predicted protein sequences derived from the following genes: atp1, 4, 6, 8, 9, cob, cox1, 2, 3, mttB, nad1, 2, 3, 4, 4L, 5, 6, 7, 9, rpl5, 6, 16, rps2, 3, 4, 7, 10, 11, 12, 13, 14, and 19. Following alignment of the sequences of individual proteins with Muscle v3.7 (Edgar Citation2004), ambiguously aligned regions were removed using TrimAL v1.4 (Capella-Gutierrez et al. Citation2009) with the options block = 6, gt = 0.7, st = 0.005, and sw = 3, and the protein alignments were concatenated using Phyutility v2.2.6 (Smith and Dunn Citation2008). For the phylogenetic analysis, the data set was partitioned by protein and the GTR + Γ4 model was applied to each of the 32 partitions.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O. 2012. Phylogeny and molecular evolution of the green algae. CRC Crit Rev Plant Sci. 31(1):1–46.
- Massjuk NP. 2006. Chlorodendrophyceae class. nov. (Chlorophyta, Viridiplantae) in the Ukrainian flora: I. The volume, phylogenetic relations and taxonomical status. Ukr Bot J. 63:601–614.
- Pombert JF, Otis C, Turmel M, Lemieux C. 2013. The mitochondrial genome of the prasinophyte Prasinoderma coloniale reveals two trans-spliced group I introns in the large subunit rRNA gene. PLoS One. 8(12):e84325.
- Robbens S, Derelle E, Ferraz C, Wuyts J, Moreau H, Van de Peer Y. 2007. The complete chloroplast and mitochondrial DNA sequence of Ostreococcus tauri: organelle genomes of the smallest eukaryote are examples of compaction. Mol Biol Evol. 24(4):956–968.
- Satjarak A, Burns JA, Kim E, Graham LE. 2017. Complete mitochondrial genomes of prasinophyte algae Pyramimonas parkeae and Cymbomonas tetramitiformis. J Phycol. 53(3):601–615.
- Smith SA, Dunn CW. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Turmel M, de Cambiaire JC, Otis C, Lemieux C. 2016. Distinctive architecture of the chloroplast genome in the chlorodendrophycean green algae Scherffelia dubia and Tetraselmis sp. CCMP 881. PLoS One. 11(2):e0148934.
- Worden AZ, Lee JH, Mock T, Rouze P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 324(5924):268–272.
