Abstract
The complete mitochondrial genome of mangrove associated plant: Sesuvium portulacastrum was analyzed in this paper, which is the first for the genus within the family Aizoaceae. The mitogenome sequence is 392,221 bp in length containing six ribosomal RNA genes, 27 transfer RNA genes, and 36 protein-coding genes. Gene nad1, nad2 and nad5 are the trans-splicing genes. One intron is found in gene ccmFc, two introns are found in genes nad4 and rps3, and four introns are found in gene nad7. Phylogenetic analysis using the maximum likelihood method positioned S. portulacastrum within the monophyletic clades of the family Aizoaceae.
Sesuvium portulacastrum belonging to Aizoaceae species, is a perennial, dichotomous, dicotyledonous halophyte plant, and often grows in coastal and inland sandy soils around the world (Yi et al. Citation2014; Chang et al. Citation2016). The species has the ability to be propagated by salt-tolerant vegetative fragments as well as to its tolerance of salt spray, sand scouring and burial, high substrate temperatures (Rabhi et al. Citation2010). It was found to be efficient in bio reclamation of salt – affected soils as a result of its aptitude to produce high biomass and to accumulate enormous sodium quantities within its shoots (Munns Citation2002). Mitochondrial genome based phylogenetic analysis would improve our understanding of the evolutionary relationship of this plant under tidal habitat. In this study, we sequenced and analyzed the complete mitochondrial DNA sequence of S. portulacastrum. This is the first complete mitogenome within the family Aizoaceae.
Fresh leaves were collected from three individual of S. portulacastrum in XinYing Mangrove Natural Garden, Hainan Island (N19°30′, E109°30′), China. The total genomic DNA was extracted from ten mixed fresh leaves of S. portulacastrum by using the modified CTAB method (Doyle Citation1987) in the laboratory of Lingnan Normal University. Further, the specimen was also stored in the herbarium of Lingnan Normal University (No. Sp20190624-001). Genome sequencing was performed on an Illumina Hiseq X Ten platform with paired-end reads of 150 bp. In total, 47.8 Gb short sequence data with Q20 was 97.16% was obtained. The remaining high-quality reads were used to assemble the mitogenome using NOVOPlasty (Dierckxsens et al. Citation2017), where Beta macrocarpa (GenBank accession FQ378026.1) used as the seed sequence. The genes annotation in the mitogenome was doing with SPAdes v.3.9.0 (Bankevich et al. Citation2012) and some genes were annotated manually. The accession number in Genbank is MN683736. Eight mitogenome sequences in Caryophyllales were aligned including S. portulacastrum, and Arabidopsis thaliana was used as the out-group species. Phylogenetic analysis using the maximum likelihood algorithm was conducted with Mega X (Kumar et al. Citation2018).
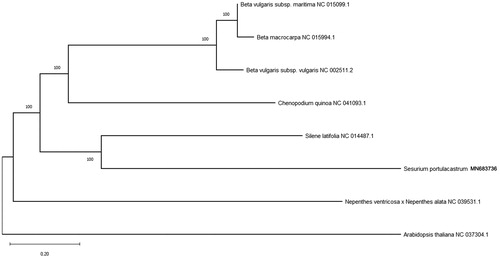
The mitogenome of S. portulacastrum is 392,221bp in length with GC content of 42.17%, which contains six ribosomal RNA genes (2 rrn16, 2 rrn12 and 2 rrn5), 27 transfer RNA genes, and 36 protein-coding genes. Among those genes, ccmFc contains one intron, nad4 and rps3 contain two introns, and nad7 contain four introns. Genes nad1, nad2, and nad5 are trans-splicing genes. Phylogenetic analysis with other seven plant mitogenomes showed that S. portulacastrum is sister to a clade containing four species in Caryophyllales (). The useful genomic resources for characterization of genetic diversity of S. portulacastrum by the mitogenome will help for the study of evolution mechanism.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski A D, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chang W, Liu X, Zhu J, Fan W, Zhang Z. 2016. An aquaporin gene from halophyte Sesuvium portulacastrum, SpAQP1, increases salt tolerance in transgenic tobacco. Plant Cell Rep. 35(2):385–395.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45(4):e18.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Munns R. 2002. Comparative physiology of salt and water stress. Plant Cell Environ. 25(2):239–250.
- Rabhi M, Giuntini D, Castagna A, Remorini D, Baldan B, Smaoui A, Abdelly C, Ranieri A. 2010. Sesuvium portulacastrum maintains adequate gas exchange, pigment composition, and thylakoid proteins under moderate and high salinity. J Plant Physiol. 167(16):1336–1341.
- Yi X, Sun Y, Yang Q, Guo A, Chang L, Wang D, Tong Z, Jin X, Wang L, Yu J, et al. 2014. Quantitative proteomics of Sesuvium portulacastrum leaves revealed that ion transportation by V-ATPase and sugar accumulation in chloroplast played crucial roles in halophyte salt tolerance. J Proteomics. 99:84–100.