Abstract
The mitochondrial genome of one leafhopper species Bolanusoides shaanxiensis was sequenced and annotated. The mitogenome is 15,724 bp in length, containing 37 typical genes and a control region. The A + T content of the whole mitogenome is 78.9%. Most of PCGs started with ATN and stopped with TAA, except for ATP8 started with TTG, COX2, COX3 and ND5 used incomplete T as stop codon. The phylogeny tree is monophyletic among 31 related species. The relationships of B. shaanxiensis and Typhlocyba sp. were closer than others. This study further enriched mitogenome database of the tribe Typhlocybini.
The genus of Bolanusoides was established by Distant (Citation1918), which belonged to the tribe Typhlocybini of the subfamily Typhlocybinae (Hemiptera: Cicadellidae) and included 13 species (Yan Citation2019). Among of them, two groups were categorized: B. heros group and B. bohater group (Dworakowska Citation1993). The species of B. shaanxiensis was described as a new species from China, which is belonged to B. heros group (Huang and Zhang Citation2005).
A male adult of B. shaanxiensis was collected from Dashahe Nature Reserve, Guizhou, China (107°61.4′E, 29°15.8′N), in September 2018. Total DNA was extracted from entire body without its abdomen using the DNeasy Blood and Tissue Kit (Cat. No. 69504). And voucher specimen’s genome DNA and male genitalia were deposited in the Institute of Entomology of Guizhou University, Guiyang, China (GUGC), accession number of them is GUGC-IDT-00188 (Zhang Citation2018). Then, the mitochondrial genome (mitogenome) of B. shaanxiensis was sequenced by Illumina NovaSeq6000 platform (Berry Genomics, Beijing, China). The reads were assembled and annotated using Generous Prime (v2019.1.3.). All tRNA genes were identified by ARWEN v1.2 (Laslett and Canbäck Citation2008). The annotated sequences of mitogenome were submitted to GenBank with accession number MN661136. All protein condoning genes (PCGs) were aligned using MAFFT algorithm in the TranslatorX (Katoh et al. Citation2017), and then poorly aligned results were removed by Gblocks 9.1 b (Castresana Citation2000; Abascal et al. Citation2010). Phylogenetic tree was reconstructed based on the 1st and 2nd codon positions of 13 PCGs using the GTR + I + G model determined by MrBayes3.2.7. on Cipres platform among B. shaanxiensis and 30 reference species.
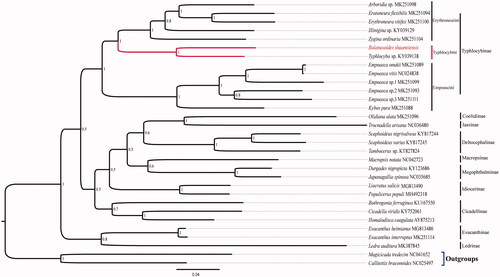
Mitogenome of B. shaanxiensis has 15,724 bp in length, and including 37 typical genes (13 PCGs, 22 tRNA genes and 2 rRNA genes), and a control region. The A + T content of genes are 78.9%, which is similar to other typhlocybine leafhopper mitogenomes (Liu et al. Citation2017; Song et al. Citation2017, Citation2019; Zhou et al. Citation2016). Most of PCGs used standardized start codon ATN and stop codon TAA, except for ATP8 started with TTG, and COX2, COX3 and ND5 genes used incomplete T as stop codon. The length of 22 tRNA is range from 61 bp (tRNA-A) to 72 bp (tRNA-K and tRNA-V). Genes of 16S rRNA and 12S rRNA are 1,174 bp and 789 bp, respectively. The results of phylogeny confirmed that relationship of selected taxon categories was monophyletic. The species of B. shaanxiensis and Typhlocyba sp. were clustered into one clade, indicating closest relationship. This work further enriched mitogenome database of the tribe Typhlocybini, with facilitating future studies on Taxonomy and Molecular systematics ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequence guided by amino acid translations. Nucleic Acids Res. 38(suppl_2):W7–W13.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Distant WL. 1918. Rhynchota. Homoptera: Appendix. Heteroptera: Addenda. The fauna of British India including Ceylon and Burma. Rhynchota. 7:2–210.
- Dworakowska I. 1993. Contribution to the knowledge of Bolanusoides Dist. (Auchenorrhyncha, Cicadellidae, Typhlocybinae, Typhlocybini). Bulletin del’ Institute Royal des Sciences Naturelles de Belgique. Entomologie. 63:59–69.
- Huang M, Zhang YL. 2005. Two new leafhopper species of Bolanusoides distant (Hemiptera: Cicadellidae: Typhlocybinae: Typhlocybini) from China. Proc Entomol Soc Wash. 107(2):428–431.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequence. Bioinform. 24(2):172–175.
- Liu HJ, Sun CY, Long J, Guo JJ. 2017. Complete mitogenome of tea green leafhopper, Empoasca onukii (Hemiptera: Cicadellidae) from Anshun, Guizhou Province in China. Mitochondrial DNA Part B Res. 2(2):808–809.
- Song N, Cai WZ, Li H. 2017. Deep-level phylogeny of Cicadomorpha inferred from mitochondrial genomes sequenced by NGS. Scientific Report. 7(1):10429:1–11.
- Song N, Zhang H, Zhao T. 2019. Insights into the phylogeny of Hemiptera from increased mitogenomic taxon sampling. Mol Phylogenet Evol. 137(2019):236–249.
- Yan B. 2019. Study on taxonomy and molecular systematics of Typhlocybini from China (Hemiptera: Cicadellidae: Typhlocybinae). China: Guizhou University; 436 pp.
- Zhang ZQ. 2018. Repositories for mite and tick specimens: acronyms and their nomenclature. Systematic & Applied Acarology. 23(12):2432 .
- Zhou NN, Wang MX, Cui L, Chen XX, Han BY. 2016. Complete mitochondrial genome of Empoasca vitis (Hemiptera: Cicadellidae). Mitochondrial DNA Part A. 27(2):1052–1053.