Abstract
In this study, we report the first complete mitochondrial genome sequence of the Aquatic Coralsnake Micrurus surinamensis. The mitochondrial genome lengthis 17,375 bp, comprising 13 protein-coding genes, 2 rRNA (12S and 16S) and 22 tRNA, as well as two typical control regions. Phylogenetic analysis based upon 13 protein-coding genes showed clusters based on terrestrial and marine species.
Keywords:
Micrurus is a genus of New World coralsnakes that includes 84 species at present. Micrurus surinamensis is the only aquatic species, with a distinct venom composition owing to its peculiar diet of fish (Aird and Silva Jr Citation2016; Silva Jr et al. Citation2018). The venom is rich in three-finger toxins (3FTxs) and presents high neurotoxic activity (Aird et al. Citation2017).
From a specimen of M. surinamensis of Altamira (Pará – Brazil; 3°20′14ʺN and 51°48′16ʺE) collected on 9 June 2014, we extracted total DNA using the chloroform–phenol protocol (Sambrook et al. Citation1989). The specimen is deposited in the herpetological collection of the Center for Biological Studies and Research of PUC-Goiás (CEPB 8984). Sequencing was performed using the Illumina MiSeq platform (309 × 2-bp reads). A paired-end library was prepared using a TruSeq DNA PCR-Free LT Sample Prep Kit (Illumina). The assembly was constructed using NOVOPlasty in ’mitochondrial’ mode (Dierckxsens et al. Citation2017). Genome annotation was performed using MITOS webserver (Bernt et al. Citation2013) and the phylogenetic analysis was performed using 13 mitochondrial protein-coding genes. These genes were aligned individually using MAFFT v. 7 (Katoh and Standley Citation2013). Alignment files were concatenated into a single matrix using Sequence Matrix software (Vaidya et al. Citation2011). Informative sites were extracted from this matrix using GBlocks v. 0.91 (Cruickshank Citation2000) and were used as input for jModelTest v. 2.1.10 (Darriba et al. Citation2012) to test the best-fitting evolutionary model for phylogeny estimation, based on the Akaike Information Criterion (AIC). Bayesian inference approach using MrBayes v. 3.2 software with 30,000,000 generations in MCMC runs was analyzed (Huelsenbeck and Ronquist Citation2001; Darling et al. Citation2012). Runs were evaluated using Tracer software (tree.bio.ed.ac.uk).
The M. surinamensis mitochondrial genome comprises 17,375 bp. It consists of 13 protein-coding genes, 2 rRNA (12S and 16S), and 22 tRNA genes, and two control regions typical of snake mitochondrial genomes. This genome has a composition of 27.5% A, 33.1% T, 27.5% G, and 12.0% C. A phylogenetic analysis was performed based on available sequences in GenBank, using the Alligator mississippiensis mitochondrial genome as an outgroup (). Three clades (Bungarus, Micrurus and Naja) represented terrestrial elapids and Laticauda exemplified aquatic elapids. Among terrestrial elapids Bungarus is a sister group of Micrurus with Naja as an outgroup, all with highly supported nodes, and patterns identical to those reported previously (Kim et al. Citation2018; Yi et al. Citation2019). The complete mitochondrial genome sequence of Micrurus surinamensis has been deposited in GenBank (accession number: MN587874).
Figure 1. Bayesian phylogenetic tree constructed using 13 protein-coding genes from complete mitochondrial genomes. Alligator mississippiensis was used as outgroup. Genbank accession numbers: Laticauda colubrina (KY496324.1), Laticauda laticaudata (KY496323.1), Laticauda semifasciata (KY496325.1), Bungarus fasciatus (EU579523.1), Bungarus multicinctus (EU579522.1), Micrurus fulvius (GU045453.1), Micrurus surinamensis (this work), Naja atra (EU913475.1), Naja naja (DQ343648.1), and Alligator mississippiensis (AKHW00000000.3).
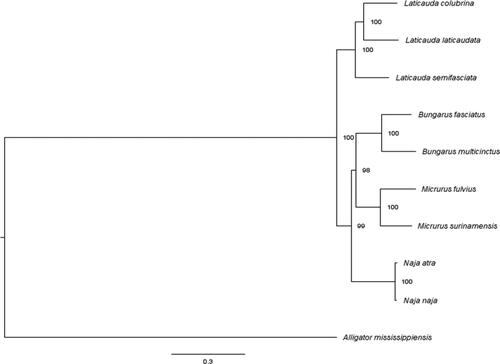
Based upon mitochondrial genomes, Micrurus surinamensis is clustered with Micrurus fulvius, as predicted they represent two different but related clades among New World coralsnakes (Roze Citation1996; Silva Jr and Sites Citation2001; Silva Jr et al. Citation2016).
Acknowledgements
We thank ‘Rede Genética Geográfica e Planejamento Regional para Conservação de Recursos Naturais no Cerrado’ (GENPAC), especially GENPAC 4 under the project ‘Modelos de nicho ecológico, distribuição potencial e o efeito de mudanças climáticas em espécies do Cerrado', and GENPAC 14, under the project ‘Filogeografia e diversidade toxinológica de Micrurus (Serpentes, Elapidae) na área de contato Cerrado-Amazônia – Caatinga’ (calling MCT/CNPq/FNDCT/FAPs/MEC/CAPES/PRO-CENTRO-OESTE no. 031/2010).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aird SD, Silva NJ Jr 2016. Química dos venenos de cobras-corais. In: Silva NJ Jr, editor. As cobras-corais do Brasil. Biologia, taxonomia, venenos e envenenamentos. Goiânia: Editora PUC Goiás; p. 242–301.
- Aird SD, Silva NJ Jr, Qiu L, Villar-Briones A, Saddi VA, Telles MPC, Grau M, Mikheyev AS. 2017. Coralsnake venomics: analyses of venom gland transcriptomes and proteomes of six Brazilian taxa. Toxins (Basel). 9(6):187.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cruickshank R. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Darling A, Ronquist F, Ayres DL, Larget B, Liu L, Teslenko M, Suchard MA, Huelsenbeck JP, Höhna S, van der Mark P. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Huelsenbeck JP, Ronquist F. 2001. MrBAYES: bayesian inference for phylogeny. Bioinformatics. 17(8):754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim IH, Park J, Suk HY, Bae HG, Min MS, Tsai TS, Park D. 2018. Phylogenetic relationships of three representative sea krait species (genus Laticauda; Elapidae; Serpentes) based on 13 mitochondrial genes. Mitochondrial DNA A. 29(5):772–777.
- Roze JA. 1996. Coral snakes of the Americas: biology, identification, and venoms. Malabar (FL): Krieger Publishing Company.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press.
- Silva FM, Prudente ALC, Machado FA, Santos MM, Zaher H, Hingst-Zaher E. 2018. Aquatic adaptations in a Neotropical coral snake: A study of morphological convergence. J Zool Syst Evol Res. 56(3):382–394.
- Silva NJ Jr, Pires MG, Feitosa DT. 2016. Diversidade das Cobras-Corais do Brasil. In: Silva NJ Jr, editor. As Cobras-Corais do Brasil: biologia, taxonomia, venenos e envenenamentos., Goiânia (Brazil): Editora PUC Goiás; p. 78–167
- Silva NJ Jr, Sites JW. 2001. Phylogeny of South American triad coral snakes (Elapidae: Micrurus) based on molecular characters. Herpetologica. 57:1–22.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Yi CH, Park J, Sasai T, Kim HS, Kim JG, Kim MS, Cho IY, Kim IH. 2019. Complete mitochondrial genome of the Ijima's Sea Snake (Emydocephalusijimae) (Squamata, Elapidae). Mitochondrial DNA Part B. 4(2):2658–2659.
