Abstract
Rotunda rotundapex (Miyata & Kishida, 1990) is a silk moth identified in Korea. We completed its mitochondrial genome which is 15,298 bp long and the shortest mitogenome of Bombycidae s.str. It includes 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a control region. Comparison with COI sequence of Taiwan isolate suggests the Korean population of this species can be a novel species. Gene order of R. rotundapex mitogenome is conserved as in other Bombycidae species. Phylogenetic trees show that R. rotundapex is closely related to genus Rondotia.
Rotunda rotundapex (Miyata & Kishida, 1990) is an East Asian silk moth named after its rounded wings. The species was first recorded in Taiwan as Bombyx rotundapex and later found in mainland China and Myanmar (Wang et al. Citation2015). In Korea, it was recorded as Bombyx shini, a novel sister species of B. rotundapex, based on its morphologies (Park and Sohn Citation2002). Despite the slight differences, a monograph on China’s silk moths treated B. shini as a junior synonym of R. rotundapex, even suggesting that the Korean population of B. shini was recently introduced by human activities (Wang et al. Citation2015). To determine its genetic background, we completed the mitochondrial genome of R. rotundapex collected in Korea.
Total DNA of R. rotundapex was extracted from a specimen collected in Hwacheon-gun, Gangwon-do, Republic of Korea (38°06′27.1″N, 127°43′26.6″E) using DNeasy Blood &Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from HiSeqX at Macrogen Inc., Korea, were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014). De novo assembly and confirmation were conducted by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li et al. Citation2009), and SAMtools 1.9 (Li Citation2013). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) and ARWEN (Laslett and Canbäck Citation2008) were used for annotation based on other silk moth mitogenomes. DNA sample and specimen (95% ethanol) were deposited in InfoBoss Cyber Herbarium (IN; J. Park, KFDS00166).
R. rotundapex (Genbank accession is MN698791) mitogenome is 15,298 bp long, the shortest in Bombycidae s.str., consisting of 13 protein-coding genes (PCGs), 2 rRNAs, 22 tRNAs, and a control region. Its GC ratio is 20.8%. Gene order is conserved in all species of Bombycoidea as in most Lepidopterans.
Partial COI gene sequence (1,459 bp) of the Taiwanese isolate (MH817447; Lin et al. Citation2019) presents 80 SNPs (5.5%) against that from our mitogenome. This divergence suggests that R. rotundapex in Korea should be considered an independent species from the Taiwanese isolate. Additional sequences of Chinese mainland’s isolates can answer the question whether the Korean population was recently introduced or not.
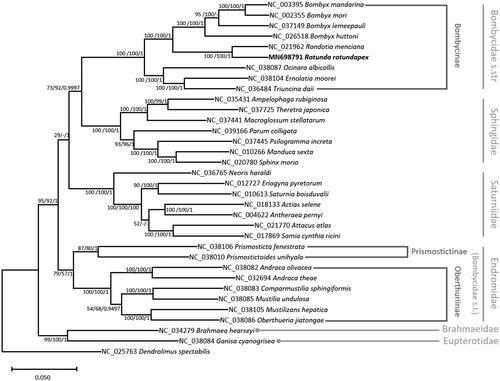
Thirteen PCGs and two rRNA genes from all available mitogenome of species in Bombycidae s.l., all available Bombycoidea genera, and an outgroup species were aligned by MAFFT 7.450 (Katoh and Standley Citation2013) and concatenated. Bootstrapped maximum likelihood, neighbor joining, and Bayesian inference trees were constructed using MEGA X (Kumar et al. Citation2018) and Mr. Bayes 3.2.6 (Huelsenbeck and Ronquist Citation2001), respectively. Our trees shows that R. rotundapex is sister to Rondotia, not Bombyx (), which is incongruent to previous study using COI and four nuclear markers (Lin et al. Citation2019). It also shows that subfamilies Oberthuriinae and Prismostictinae are far from Bombycinae (), agreeing that former subfamilies should be treated under Endromidae (Wang et al. Citation2019). Sphingidae, however, is clustered with Bombycidae s.str not Saturniidae (), which is also incongruent to former phylogenomic study (Hamilton et al. Citation2019) with relatively low bootstrap values and posterior priority (). Our results address the question why trees of mitochondrial and nucleic genes present difference phylogenetic relationships at the level of higher taxa.
Figure 1. Maximum likelihood (bootstrap repeat is 1,000), neighbor joining (bootstrap repeat is 10,000), and Bayesian inference (1,000,000 generations) phylogenetic trees of all available silk moth (Bombycidae s.l.) mitochondrial genomes: Rotunda rotundapex (MN698791 in this study), Bombyx mandarina (NC_003395), Bombyx mori (NC_002355), Bombyx lemeepauli (NC_037149), Bombyx huttoni (NC_026518), Rondotia menciana (NC_021962), Ocinara albicollis (NC_038087), Ernolatia moorei (NC_038104), Triuncina daii (NC_036484), Prismosticta fenestrata (NC_038106), Prismostictoides unihyala (NC_038010), Andraca olivacea (NC_038082), Andraca theae (NC_032694), Comparmustilia sphingiformis (NC_038083), Mustilia undulosa (NC_038085), Mustilizans hepatica (NC_038105), Oberthueria jiatongae (NC_038086), as well as 16 moths of all available Bombycoidea genera: Ampelophaga rubiginosa (NC 035431), Theretra japonica (NC 037725), Macroglossum stellatarum (NC_037441), Parum colligata (NC_039166), Psilogramma increta (NC_037445), Manduca sexta (NC_010266), Sphinx morio (NC_020780), Neoris haraldi (NC_036765), Eriogyna pyretorum (NC_012727), Saturnia boisduvalii (NC_010613), Actias selene (NC_018133), Antheraea pernyi (NC_004622), Attacus atlas (NC_021770), Samia cynthia ricini (NC_017869), Brahmaea hearseyi (NC_034279), Ganisa cyanogrisea (NC_038084), and a lappet moth: Dendrolimus spectabilis (NC_025763) as an outgroup. Phylogenetic tree was drawn based on maximum likelihood tree. The numbers above branches indicate bootstrap support values of maximum likelihood, neighbor joining trees, and posterior probability of Bayesian inference tree, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Hamilton CA, St Laurent RA, Dexter K, Kitching IJ, Breinholt JW, Zwick A, Timmermans MJ, Barber JR, Kawahara AY. 2019. Phylogenomics resolves major relationships and reveals significant diversification rate shifts in the evolution of silk moths and relatives. BMC Evol Biol. 19(1):1–13.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Lin R-J, Braby MF, Hsu Y-F. 2019. The immature stages, biology, and phylogenetic relationships of Rotunda rotundapex (Lepidoptera: Bombycidae). J Insect Sci. 19(2):19.
- Park K-T, Sohn J-C. 2002. Description of Bombyx shini sp. nov. (Lepidoptera, Bombycidae) from Korea. Tinea. 17(2):67–69.
- Wang X, Chen ZM, Gu XS, Wang M, Huang GH, Zwick A. 2019. Phylogenetic relationships among Bombycidae sl (Lepidoptera) based on analyses of complete mitochondrial genomes. Syst Entomol. 44(3):490–498.
- Wang X, Wang M, Zolotuhin VV, Hirowatari T, Wu S, Huang G-H. 2015. The fauna of the family Bombycidae sensu lato (Insecta, Lepidoptera, Bombycoidea) from Mainland China, Taiwan and Hainan Islands. Zootaxa. 3989(1):1–138.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(Suppl 14):S2.
