Abstract
The complete mitochondrial genome of the Annamanum lunulatum is 15,610 bp in length, which contains 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and the A + T-rich region. The arrangement of genes is identical to all know longhorn beetles mitochondrial genomes. The overall AT content of the mitochondrial genome is 75.3%, whereas the AT content of A + T-rich region is 84.3%. In ML and BI phylogenetic analyses, A. lunulatum is a sister clade to Blepephaeus succinctor, and the monophyly of Lamiinae is supported.
Coleoptera (Hexapoda: Insecta) are a group of insects with over 360,000 described species of beetles (Hunt et al. Citation2007). It is in the larval stage that some species of Coleoptera belonging to pests make strong damage to plants. Annamanum lunulatum (Cerambycidae: Lamiinae) is a longhorn beetle firstly reported by Pic (Citation1934) and Uraecha longzhouensis is considered as one of its junior synonyms (Lin and Lingafelter Citation2018). The genus Annamanum includes 31 described species located in South China, Japan, Vietnam, Laos, Cambodia, India, Myanmar, and Malaysia, among which 15 species were reported from China (Holzschuh Citation2017; Lin and Ge Citation2017; Tavakilian and Chevillotte Citation2018; Yang and Yang Citation2019). However, there were few genetic information on the mitochondrial genome reported in this genus. Hence, we sequenced the mitochondrial genome of A. lunulatum to benefit the studies of the genetic diversity in Annamanum and to discuss its phylogenetic relationship within Lamiinae.
Samples of A. lunulatum were collected from Jinxiu (22°57′28″ N, 107°11′38″ E), Guangxi province, China on 10 August 2016 and classified by JY Zhang. The sample (JX20160810) was identified and stored at 40 °C in the Animal Specimen Museum, College of Life Sciences and Chemistry, Zhejiang Normal University, China. The total genomic DNA was extracted from leg muscle tissue using an Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China) and stored in the Zhang’s laboratory. Eight universal primers and six specific primers (Simon et al. Citation2006; Zhang et al. Citation2008; Zhang, Cai, et al. Citation2018; Zhang, Yu, et al. Citation2018) were utilized for polymerase chain reaction (PCR) amplification. The mitochondrial genome was deposited in GenBank with an accession number MN356095.
The complete mitochondrial genome of A. lunulatum is 15,610 bp in length, including 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and the A + T-rich region. The overall AT content of the mitochondrial genome is 75.3%, whereas the AT content of A + T-rich region is 84.3%. Almost all the protein-coding genes use ATN as an initiation codon excluding ND1 with TTG. Eleven genes (ND1, ND2, ND3, ND4L, COI, COII, COIII, ATP6, ATP8, ND6, Cyt b) end the typical stop codon TAA or TAG, whereas ND4 and ND5 end with T—.
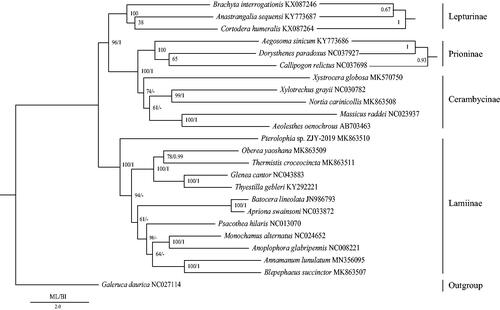
In order to reconstruct the phylogenetic relationships of Lamiinae, we used 13 protein-coding genes of 24 species, which contained Galeruca daurica as the outgroup (Kim et al. Citation2009; Lu et al. Citation2011; Chiu et al. Citation2016; Fang et al. Citation2016; Guo et al. Citation2016; Li et al. Citation2016; Wang et al. Citation2016; Lim et al. Citation2017; Liu et al. Citation2017; Yang et al. Citation2017; Liu et al. Citation2018; Que et al. Citation2019; Wang, Dai, et al. Citation2019; Wang, Lan, et al. Citation2019). MrBayes version 3.2 and the RAxML version 8 programs (Ronquist et al. Citation2012; Stamatakis Citation2014) were used to construct BI and ML trees, respectively (). Annamanum lunulatum was a sister clade to Blepephaeus, and the monophyly of Lamiinae was supported in both BI and ML analyses.
Figure 1. Phylogenetic trees of the relationships among 24 species of Coleoptera, including Annamanum lunulatum, were based on the nucleotide dataset of the 13 mitochondrial protein-coding genes. The numbers showed between branches indicate the posteriori probabilities from Bayesian inference (BI) and bootstrap percentages from maximum-likelihood (ML, 1000 replications) analyses. The GenBank accession numbers of all species were also shown.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chiu WC, Yeh WB, Chen ME, Yang MM. 2016. Complete mitochondrial genome of Aeolesthes oenochrous (Fairmaire) (Coleoptera: Cerambycidae): an endangered and colorful longhorn beetle. Mitochondrial DNA A. 27(1):686–687.
- Fang J, Qian L, Xu M, Yang XJ, Wang BD, An YL. 2016. The complete nucleotide sequence of the mitochondrial genome of the Asian longhorn beetle, Anoplophora glabripennis (Coleoptera: Cerambycidae). Mitochondrial DNA A. 27(5):3299–3300.
- Guo K, Chen J, Xu CQ, Qiao HL, Xu R, Zhao XJ. 2016. The complete mitochondrial genome of the longhorn beetle Xylotrechus grayii (Coleoptera: Cerambycidae). Mitochondrial DNA A. 27:2133–2134.
- Holzschuh C. 2017. Neue Arten von Bockkäfern aus der Tribus Clytini und der Unterfamilie Lamiinae (Coleoptera, Cerambycidae) vom asiatischen Festland. Acta Musei Moraviae. Scientiae Biologicae (Brno). 102:93–138.
- Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A, John OS, Wild R, Hammond PM, Ahrens D, Balke M, Caterino MS, et al. 2007. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 318(5858):1913–1916.
- Kim KG, Hong MY, Kim MJ, Im HH, Kim MI, Bae CH, Seo SJ, Lee SH, Kim I. 2009. Complete mitochondrial genome sequence of the yellow-spotted long-horned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol Cells. 27(4):429–441.
- Li F, Zhang H, Wang W, Weng H, Meng Z. 2016. Complete mitochondrial genome of the Japanese pine sawyer, Monochamus alternatus (Coleoptera: Cerambycidae). Mitochondrial DNA A. 27(2):1144–1145.
- Lim J, Yi DK, Kim Y H, Lee W, Kim S, Kang JH, Kim IK. 2017. Complete mitochondrial genome of Callipogon relictus Semenov (Coleoptera: Cerambycidae): a natural monument and endangered species in Korea. Mitochondrial DNA B. 2(2):629–631.
- Lin MY, Lingafelter SW. 2018. Taxonomic notes on Chinese Lamiini (Coleoptera: Cerambycidae: Lamiinae). Zootaxa. 4482(2):367–374.
- Lin MY, Ge SQ. 2017. Notes on the genera Annamanum Pic and Uraecha Thomson (Coleoptera: Cerambycidae: Lamiinae: Lamiini). Humanity Space Int Almanac. 6:889–915.
- Liu JH, Jia PF, Luo T, Wang QM. 2017. Complete mitochondrial genome of white-striped long-horned beetle, Batocera lineolata (Coleoptera: Cerambycidae) by next-generation sequencing and its phylogenetic relationship within superfamily Chrysomeloidea. Mitochondrial DNA B. 2(2):520–521.
- Liu YQ, Chen DB, Liu HH, Hu HL, Bian HX, Zhang RS, Yang RS, Jiang XF, Shi SL. 2018. The complete mitochondrial genome of the longhorn beetle Dorysthenes Paradoxus (Coleoptera: Cerambycidae: Prionini) and the implication for the phylogenetic relationships of the Cerambycidae species. J Insect Sci. 18(2):21:1–8.
- Lu W, Wang Q, Tian MY, Xu J, Qin AZ. 2011. Phenology and laboratory rearing procedures of an Asian longicorn beetle, Glenea cantor (Coleoptera: Cerambycidae: Lamiinae). J Econ Entomol. 104(2):509–516.
- Pic M. 1934. Nouveautés asiatiques. Matériaux Pour Servir à L’étude Des Longicornes. 11:33–40.
- Que S, Yu A, Liu P, Jin M, Xie GA. 2019. The complete mitochondrial genome of Apriona swainsoni. Mitochondrial DNA B. 4(1):931–932.
- Ronquist F, Teslenko M, Mark PVD, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37(1):545–579.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tavakilian G, Chevillotte H. 2018. Titan: base de données internationales sur les Cerambycidae ou Longicornes. Version 4.0. [accessed June 2019]. http://titan.gbif.fr/index.html.
- Wang J, Dai XY, Xu XD, Zhang ZY, Yu DN, Storey KB, Zhang JY. 2019. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: Cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ. 7:e7633.
- Wang J, Lan DY, Dai XY, Yu DN, Storey KB, Zhang JY. 2019. The complete mitochondrial genome of Xystrocera globose (Coleoptera: Cerambycidae) and its phylogeny. Mitochondrial DNA B. 4(1):1647–1649.
- Wang YT, Liu YX, Tong XL, Ren QP, Jiang GF. 2016. The complete mitochondrial genome of the longhorn beetle, Massicus raddei. Mitochondrial DNA A. 27(1):209–211.
- Yang J, Cao LJ, Geng YX, Wei DF, Chen M. 2017. Determination of the complete mitochondrial genome of Thyestilla gebleri and comparative analysis of the mitochondrial genome in Cerambycidae. Chinese J Appl Entomol. 54:755–766.
- Yang SL, Yang SY. 2019. Annamanum flavimaculatum, a new species of longhorn beetle (Coleoptera, Cerambycidae) from China. Zookeys. 889:49–56.
- Zhang JY, Zhou CF, Gai YH, Song DX, Zhou KY. 2008. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene. 424(1-2):18–24.
- Zhang LP, Cai YY, Yu DN, Storey KB, Zhang JY. 2018. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: Toxoderidae). PeerJ. 6:e459.
- Zhang LP, Yu DN, Storey K B, Cheng HY, Zhang JY. 2018. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea. Int J Biol Macromol. 111:787–795.
