Abstract
Fairy shrimps (Anostraca) constitute an important component of seasonally aquatic habitats, but few complete mitochondrial genomes have been published for this group. Here, we report the mitogenome of a common southern African species, Streptocephalus cafer, from Botswana (accession number: MN720104). Low-coverage shotgun sequencing recovered two contigs 15653 bp and 1347 bp in length that are separated by a repetitive region of unknown length within the non-coding control region. The mitogenome’s GC content is 31.80%. Phylogenetic analysis using protein-coding genes confirms the sister taxon relationship of S. cafer with the only other congener whose mitogenome has been reconstructed to date, the Asian S. sirindhornae.
Fairy shrimps (Anostraca) are freshwater crustaceans that occur exclusively in seasonally astatic aquatic habitats (Brendonck et al. Citation2008; Rogers Citation2009). Similar to other branchiopod members of the order, fairy shrimps produce dormant eggs which allow the survival of the next generation through dry periods (Brendonck et al. Citation2008; Rogers Citation2009) and demonstrate rapid body growth rates to facilitate attainment of sexual maturity within limited hydroperiod windows (Rogers Citation2009). The Anostraca originated in the Cambrian (Harvey et al. Citation2012), with the monobasic Streptocephalidae probably appearing during the early Cretaceous, approximately 105 million years ago (Daniels et al. Citation2004). The genus Streptocephalus has about 66 species worldwide, with numerous species occuring in southern Africa (Rogers Citation2013; Rogers and Padhye Citation2014; Shu et al. Citation2018). Despite their ecological significance, only a few complete mitogenomes of Anostracans have been published (Fan et al. Citation2015; Liu et al. Citation2016). Here, we describe the first complete genome of a widespread southern African fairy shrimp, Streptocephalus cafer (Lovén, 1847) (sensu Hamer et al. Citation1994).
Specimens of S. cafer were collected from a temporary pool on the outskirts of Palapye, Central District, Botswana (27.16616 E, 22.54507 S) and stored separately in 1.5 mL tubes containing 80% ethanol. Voucher specimens from the locality were lodged at the Kansas Biological Survey (DCR-1136). Total genomic DNA was extracted using the CTAB method (Doyle & Doyle 1987). One µg of DNA was used to prepare a genomic DNA library using the NEBNext DNA Library Preparation Kit (Massachusetts, USA). The library was then sequenced on an Illumina Hi-Seq platform using 2 × 150 chemistry with an average insert length of 350 bp.
The sequencing run yielded 59,848,500 paired-end sequences. The complete mitogenome was assembled using NOVOPlasty v3.5 (Dierckxsens et al. Citation2017) and annotated in MitoZ v.2.4 . The mitogenome assembly resulted in two contigs (15653 bp and 1347 bp, respectively) separated by a repetitive region of unknown length within the non-coding control region. MITOS annotation on the longest contig identified all 13 protein-coding genes, 22 tRNAs and 2 rRNAs, typical of crustaceans. The GC content of the total assembly was estimated at 31.8%. MitoZ annotation reported several instances of non-canonical start codons and truncated stop codons, consistent with other studies on arthropods (Monsanto et al. Citation2019).
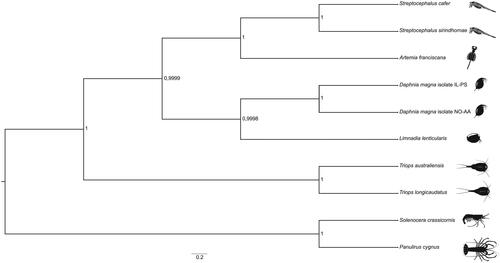
Protein-coding sequences from S. cafer and the complete mitogenomes of nine related species were aligned in MAFFT v7.429 (Katoh et al. Citation2009). A Bayesian phylogenetic tree was reconstructed with BEAST2 (Bouckaert et al. Citation2014) using default parameters, except that the substitution model was changed to HKY (Hasegawa et al. Citation1985) with four gamma categories. BEAST2 was run for 50,000,000 iterations with 30% burn-in. The convergence of the chain and Effective Sample Size (ESS) was assessed in Tracer v1.7 (Rambaut et al. Citation2018), and the resulting phylogenetic tree was visualized in FigTree v1.4 (Rambaut and Drummond Citation2012) (). It confirms the sister taxon relationship of S. cafer with the only other Streptocephalus species whose mitogenome has been reconstructed to date, the Asian S. sirindhornae.
Figure 1. A Bayesian phylogenetic tree constructed in BEAST2 using mitogenome sequences of Streptocephalus cafer (NCBI accession number MN720104) and nine other crustacean species: Streptocephalus sirindhornae (NC_026704.1), Daphnia magna isolate IL-PS (MH683649.1), Daphnia magna isolate NO-AA(MH683655.1), Artemia franciscana (X69067.1), Limnadia lenticularis (NC_039394.1), Panulirus cygnus (KT696496.1), Solenocera crassicornis (KU899137.1), Triops australiensis (LK391946.1) and Triops longicaudatus (AY639934.1). The numbers next to each node represent posterior probability and the scale bar shows the scaled substitution rate.
Acknowledgements
We acknowledge the Botswana International University of Science and Technology (BIUST), National Research Foundation (grant no. 117700), Centre for High Performance Computing, South Africa and University of Johannesburg for providing funding and facilities for the study. The Ministry of Environment, Natural Resources Conservation and Tourism (Botswana) is thanked for the issuing of a research permit (ENT 8/36/4XXXXII(14)).
Disclosure statement
No existing competition and/or financial interest has been declared by the authors. The authors declare that they own the opinions, findings, conclusions or recommendations expressed in this material.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537.
- Brendonck L, Rogers DC, Olesen J, Weeks S, Hoeh R. 2008. Global diversity of large branchiopods (Crustacea: Branchiopoda) in fresh water. Hydrobiologia. 595(1):167–176.
- Daniels SR, Hamer M, Rogers C. 2004. Molecular evidence suggests an ancient radiation for the fairy shrimp genus Streptocephalus (Branchiopoda: Anostraca). Biol J Linn Soc. 82(3):313–327.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18
- Fan YP, Lu B, Yang JS. 2015. The complete mitogenome of the fairy shrimp Phallocryptus tserensodnomi (Crustacea: Anostraca: Thamnocephalidae). Mitochondrial DNA. Part A. DNA Mapping, Sequencing, and Analysis. 27:3313–3314.
- Hamer M, Brendonck L, Coomans A, Appleton C. 1994. A review of the African Streptocephalidae (Crustacea: Branchiopoda: Anostraca) Part 1: south of Zambezi and Kunene rivers. Arch Hydrobiol. 99:235–277.
- Harvey THP, Velez MI, Butterfield NJ. 2012. Exceptionally preserved crustaceans from western Canada reveal a cryptic Cambrian radiation. P Natl Acad Sci USA. 109(5):1589–1594.
- Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22(2):160–174.
- Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 537:39–64.
- Liu XC, Li HW, Jermnak U, Yang JS. 2016. The complete mitogenome of the freshwater fairy shrimp Streptocephalus sirindhornae (Crustacea: Anostraca: Streptocephalidae). Mitochondrial DNA. Part A. 27:3189–3191.
- Monsanto DM, Jansen van Vuuren B, Jagatap H, Jooste CM, Janion-Scheepers C, Teske PR, Emami-Khoyi A. 2019. The complete mitogenome of the springtail Cryptopygus antarcticus travei provides evidence for speciation in the Sub-Antarctic region. Mitochondrial DNA Part B. 4(1):1195–1197.
- Rambaut A, Drummond A. 2012. FigTree: tree figure drawing tool, v1. 4.2. Scotland: Institute of Evolutionary Biology, University of Edinburgh.
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. System Biol. 67(5):901–904.
- Rogers DC. 2009. Branchiopoda (Anostraca, Notostraca, Laevicaudata, Spinicaudata, Cyclestherida). In: Likens, G. F. editor, Encyclopedia of Inland Waters. Cambridge, MA: Academic. Press; Vol. 2. p. 242–249.
- Rogers D.C. 2013. Anostraca catalogus (Crustacea: Branchiopoda). Raffles B Zool. 61:525–546.
- Rogers DC, Padhye S. 2014. A new species of Streptocephalus (Crustacea: Anostraca: Streptocephalidae) from the Western Ghats, India, with a key to the Asian species. Zootaxa. 3802(1):75–84.
- Shu SS, Rogers DC, Chen XY, Sanoamuang L. 2018. Streptocephalus diversity in Myanmar, with description of a new species (Branchiopoda, Anostraca). Zookeys. 734:1–12.
