Abstract
We reported the complete mitochondrial genome yielded by next-generation sequencing of Brochis multiradiatus in this study. The total length of the mitochondrial genome is 16,916 bp, with the base composition of 32.49% A, 25.47% T, 27.12% C, and 14.91% G. It contains 2 ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and a major non-coding control region (D-loop region). The arrangement of these genes is the same as that found in the Corydoras. The complete mitochondrial genomes of B. multiradiatus and other 12 species from Siluriformes were used for phylogenetic analysis using neighbor-joining method. The topology demonstrated that all species belong to four genera and are divided into two groups (Siluridae and Callichthyidae), the B. multiradiatus was clustered with genus Corydoras. Brochis multiradiatus’ molecular classification is consistent with the external morphological feature results, so the information of the mitogenome could be used for future identification of Brochis.
Brochis multiradiatus belongs to Teleostei, Siluriformes, Callichthyidae, subfamily Corydoradinae. Brochis multiradiatus is common in Chinese ornamental fish market. Because there are two small ‘whiskers’ beside its mouth, which looks like a small mouse swimming in the water, it is called mousefish. Mousefish is a category fish just named from appearance, such as Corydoras nattereri voucher (Moreira et al. Citation2016), Corydoras panda (Liu, Liu, Xiao, et al. Citation2019), Corydoras duplicareus (Liu, Xu, Xiao Citation2019), Corydoras arcuatus (Liu, Xu, Xiao, Liu Citation2019), and Corydoras sterbai (Liu, Liu, Xu, et al. Citation2019), they are all named mousefish. In order to distinguish and identify B. multiradiatus in genome, we determined the mitochondrial genome sequence of the B. multiradiatus and carried out mitochondrial genome structure and phylogenetic analysis. The live sample of B. multiradiatus was collected from the Red Star Ornamental Fish Market in Changsha, Hunan Province, China (113.03E, 28.09 N). After anesthesia with MS-222 (3-Aminobenzoic acid ethyl ester methanesulfonate), dorsal muscle tissue was collected and preserved in 99% ethanol in Museum of Hunan Agricultural University. After DNA extraction (Tissue DNA Kit D3396-02, Omega, Bio-Tek, Norcross, GA) and sequencing library construction (Sangon Biotech, Shanghai, China), paired-end reads were sequenced using HiSeq XTen PE 150 of Illumina. After extracting the DNA of the tissue (Number: QT001), the sequence library was constructed. BBduk and BLASTp were used to assess and monitor data quality. NOVOPlasty and SPAdes were used for de novo assembly. MITOS2 server and Geneious R11 (Tan et al. Citation2019) were used to predict and annotate the mitochondrial genome. Geneious Tree Builder was used for phylogenetic analysis and building phylogenetic tree.
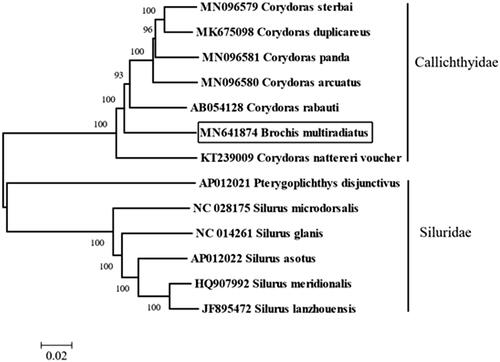
Totally 27,191,198 high-quality clean reads (150 bp PE read length) were obtained. The total length of the B. multiradiatus mitochondrial genome is 16,916 bp (GenBank accession number: MN641874), with the base composition of 32.49% A, 25.47% T, 27.12% C, and 14.91% G. It contains 2 ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and a major non-coding control region 1298 bp in length. The arrangement of these genes is the same as that found in the Siluriformes (Liu, Xu, Xiao Citation2019). All the protein initiation codons are ATG, except for cox1 that begins with GTG. The complete mitogenomes of B. multiradiatus and other 12 species from Siluriformes were used for phylogenetic analysis. The neighbor-joining tree was built by Geneious with Tamura–Nei (genetic distance) model and global aligment with free end gaps (aligment type) showed all species belong to 4 genera are divided into two groups (Siluridae and Callichthyidae), and the B. multiradiatus was clustered with other species from genus Corydoras (). Brochis multiradiatus’ external morphological feature classification is consistent with the molecular classification results, so the information of the mitogenome could be used for future phylogenetic analysis and identification of B. multiradiatus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Liu Q, Liu Y, Xiao T, Xu B. 2019. Complete mitochondrail genome of Corydoras panda (Teleostei, Siluriformes, Callichthyidae, Corydoradinae). Mitochondrial DNA Part B. 4(2):2878–2879.
- Liu Q, Liu Y, Xu B, Xiao T. 2019. Next-generation sequencing yields the complete mitochondrial genome of Corydoras sterbai (Teleostei, Siluriformes, Callichthyidae, Corydoradinae). Mitochondrial DNA Part B. 4(2):2880–2881.
- Liu Q, Xu B, Xiao T. 2019. Complete mitochondrial genome of Corydoras duplicareus (Teleostei, Siluriformes, Callichthyidae). Mitochondrial DNA Part B. 4(1):1832–1833.
- Liu Y, Xu B, Xiao T, Liu Q. 2019. Characterization and phylogenetic analysis of Corydoras arcuatus mitochondrial genome. Mitochondrial DNA Part B. 4(2):2876–2877.
- Moreira DA, Buckup PA, Britto MR, Magalhães MGP, Andrade PCCd, Furtado C, Parente TE. 2016. The complete mitochondrial genome of corydoras nattereri (callichthyidae: corydoradinae). Neotrop Ichthyol. 14(1).
- Tan J, Yin Z, Huang H, Zeng C. 2019. Characterization and phylogenetic analysis of Acheilognathus chankaensis mitochondrial genome. Mitochondrial DNA B. 4(1):1148–1149.