Abstract
Hovenia acerba Lindl. is an important medicinal plant, for which complete chloroplast genome (Accession: MN782301) was sequenced, assembled and annotated. The genome size is 161,668 bp and the overall GC content is 36.69%, with large single-copy (LSC, 89,451bp) regions, small single-copy (SSC, 18,979 bp) regions, and two inverted repeat regions (IRs, 26,619 bp each). A total of 130 genes are successfully annotated, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenetic relationships showed that H. acerba is closely related to the species of Ziziphus genus.
Hovenia acerba Lindl. is a medicinal plant, belonging to Hovenia of Rhamnaceae family (Xu et al. Citation2012). Its pedicle is fleshy and fat, sweet in taste, and edible in a mature period. It is widely cultivated in many regions of China, such as Shanxi, Sichuan, Chongqing, Zhejiang, Shandong, etc. (Liu et al. Citation2017). It has antioxidant, antineoplastic, and hangover properties, and has been used broadly in medical research (Xiang et al. Citation2011; Zhang et al. Citation2019). The complete chloroplast (cp) genome can provide valuable genomic information for the conservation and phylogenetic studies of valuable species (Liu et al. Citation2019). However, the cp genome of H. acerba Lindl. has not been fully sequenced. In this present study, we reported and characterized the complete chloroplast (cp) genome of H. acerba based on Illumina pair-end sequencing and compared it with other genus chloroplast genome sequences. It is helpful for the evolution process and genetics conservation of Hovenia genus.
The fresh leaves of H. acerba Lindl. (Voucher specimen Accession No. SDGZ0018) were collected from the Wanjishan field of Shandong Institute of Pomology (36.21°N, 117.08°E), Shandong Province, China. Total genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands). cpDNA sequencing was performed with an Illumina Hiseq 2500 platform by Nanjing Genepioneer Biotechnologies. The fastp program was used to filter the raw paired-end reads of cpDNA (Chen et al. Citation2018). The GetOrganelle was used to perform de novo assembly (Jin et al. Citation2018). The cp genome was annotated using the program DOGMA (Wyman et al. Citation2004). The OGDRAW was used to generate the circular genome map of the genome (Lohse et al. Citation2013). The complete cp genome of H. acerba was deposited in the GenBank (Accession: MN782301).
The complete cp genome of H. acerba was 161,668 bp in length with an overall GC content of 36.69%. The cp genome is made up of a large single-copy region (LSC) of 89,451 bp, small single-copy region (SSC) with 18,979bp, and two inverted repeat regions (IRs) with 26,619 bp. The whole complete cp genome encoded 130 unique genes, which contained 85 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The tRNA genes are distributed throughout the whole genome with 22 in the LSC, one in the SSC, and 14 in the IR regions, while rRNAs are only situated in the IR regions. There were 18 duplicate genes in IRs, including seven protein-coding genes (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, rps12), seven tRNA genes (trnICAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnRACG, trnN-GUU), and four rRNA genes (rrn16S, rrn23S, rrn4.5S, rrn5S). Among the protein-coding genes, three genes (clpP, rps12, ycf3) contained two introns, and other nine genes (atpF, ndhA, ndhB, petB, petD, rp116, rpl2, rpoC1, rps16) had one intron each.
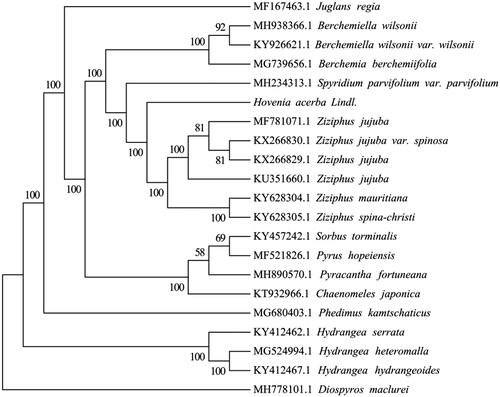
To ascertain the phylogenetic position of H. acerba, 18 complete cp genomes within Rhamnaceae family were selected, Juglans regia (GenBank Accession No. MF167463.1) and Diospyros maclurei (GenBank Accession No. MH778101.1) were considered as outgroup. The cp genomes of these species were aligned using MAFFT v7.3 (Kazutaka and Standley Citation2013). The maximum likelihood (ML) phylogenetic tree was constructed by the IQ-TREE with the best-fit model identified using ModelFinder (Kalyaanamoorthy et al. Citation2017). The result showed that H. acerba is closely related to the species of Ziziphus genus (). It will be valuable for the genetic study on the Hovenia and Ziziphus genus in Rhamnaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Jin JJ, Yu WB, Jun B, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu C, Wu JL, Zhao N, Sun J, Shen XX, Wang ZA. 2017. Correlation analysis of Hovenia acerba quality with origin and appearance traits. China J Chinese Mater Med. 24:4769–4774.
- Liu HL, Zhang Y, Zhang XN, Geng ML, Li MM. 2019. The first complete chloroplast genome of Zelkova schneideriana Hand.-Mazz. (Ulmaceae). Mitochondrial DNA B. 4(1):378–379.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome-DRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41(W1):W575–W581.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Xu F-F, Zhang X-Q, Zhang J, Liu B, Jiang J, Wang W-J, Gao M-H, Jiang R-W, Ye W-C. 2012. Two methyl-migrated 16,17-seco-dammarane triterpenoid saponins from the seeds of Hovenia acerba. J Asian Nat Prod Res. 14(2):135–140.
- Xiang JL, Li ZX, Li H, Wu QH, Zheng SY. 2011. Separation and antioxidant activity of polyphenols with different polarities from Hovenia acerba fruit. Food Sci. 32:25–29.
- Zhang Q, Liu Y, Hu HF, Wu CZ, Jiang JJ, Li C. 2019. Screening of antitumor activity ingredients in Hovenia acerba ethyl acetate. Modern Chin Med. 6:782–785.